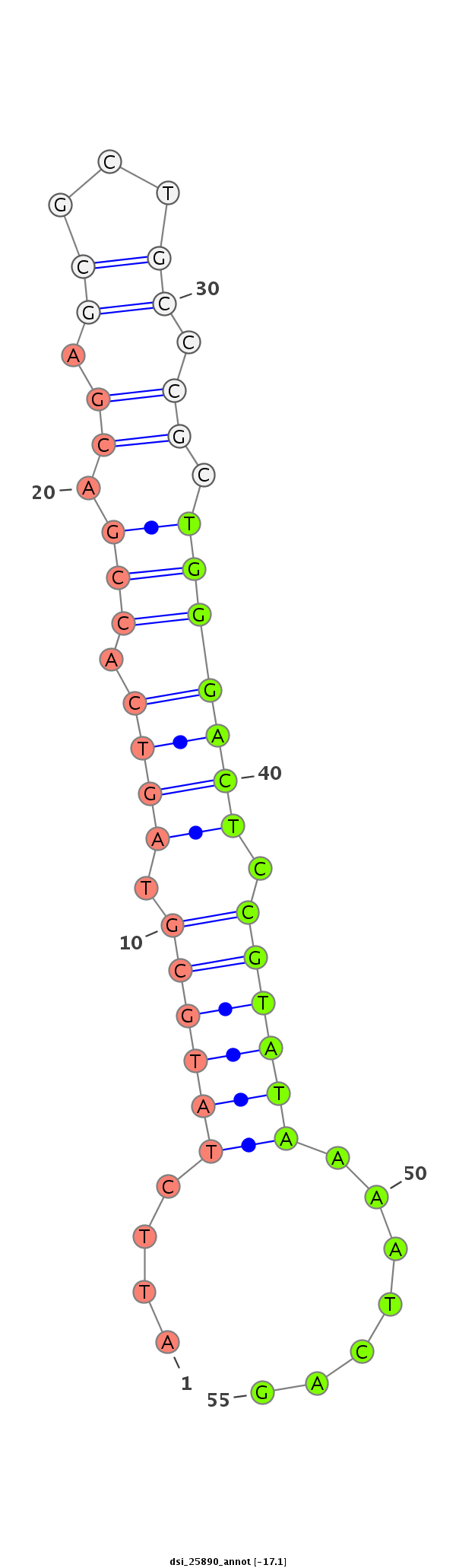
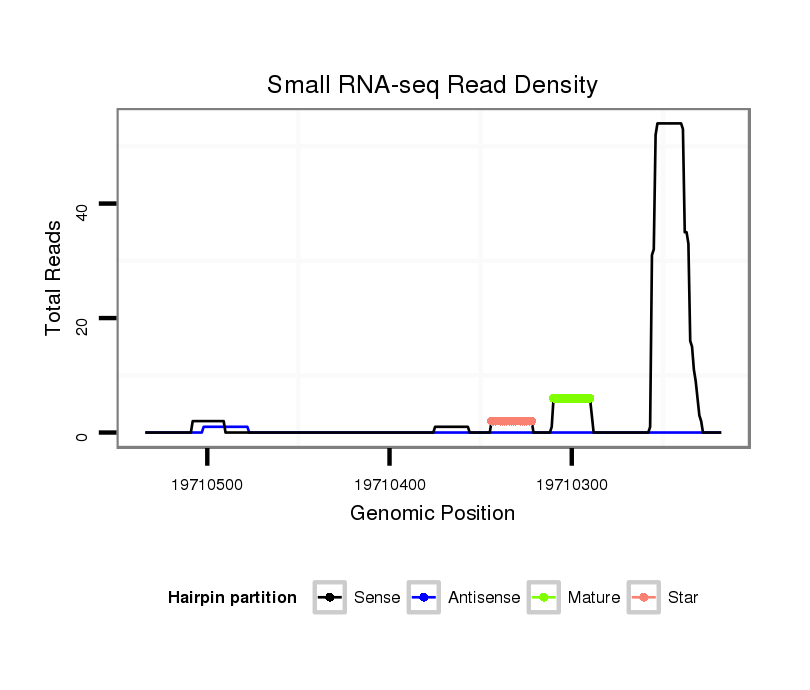
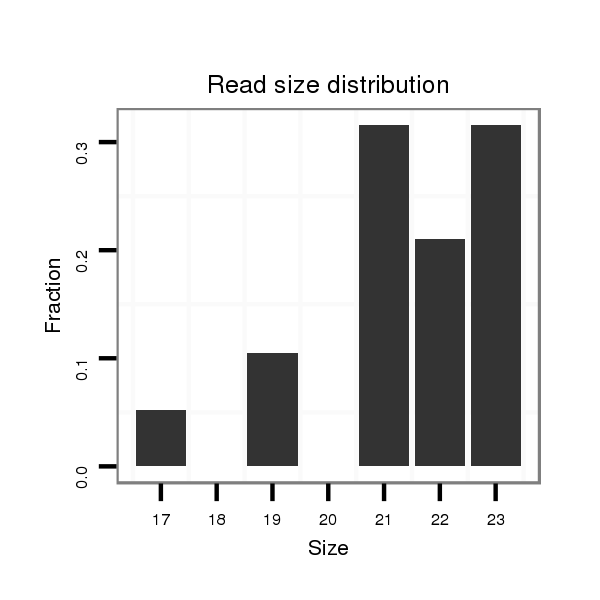
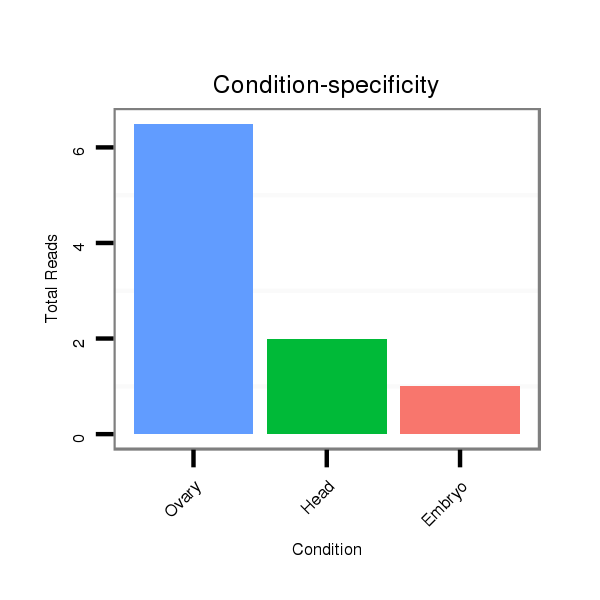
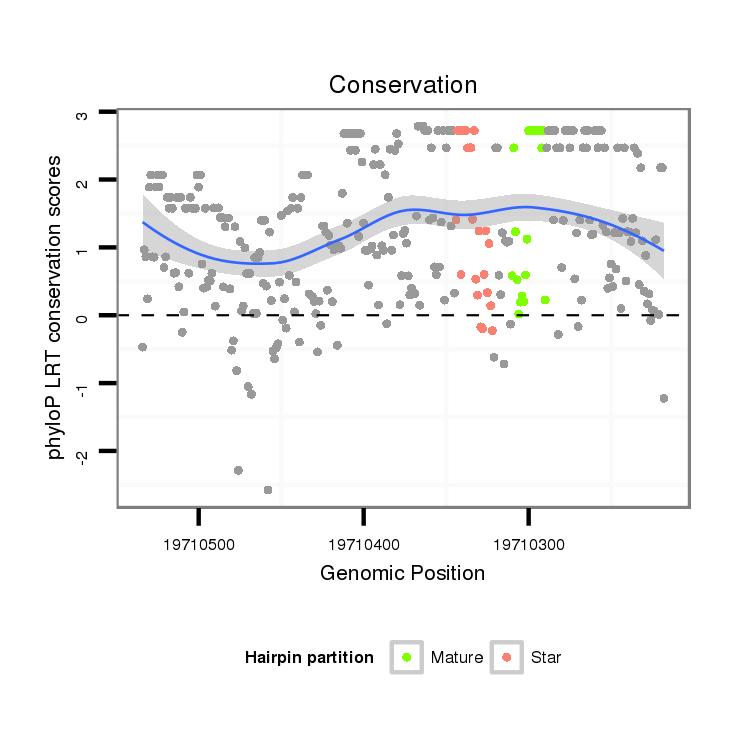
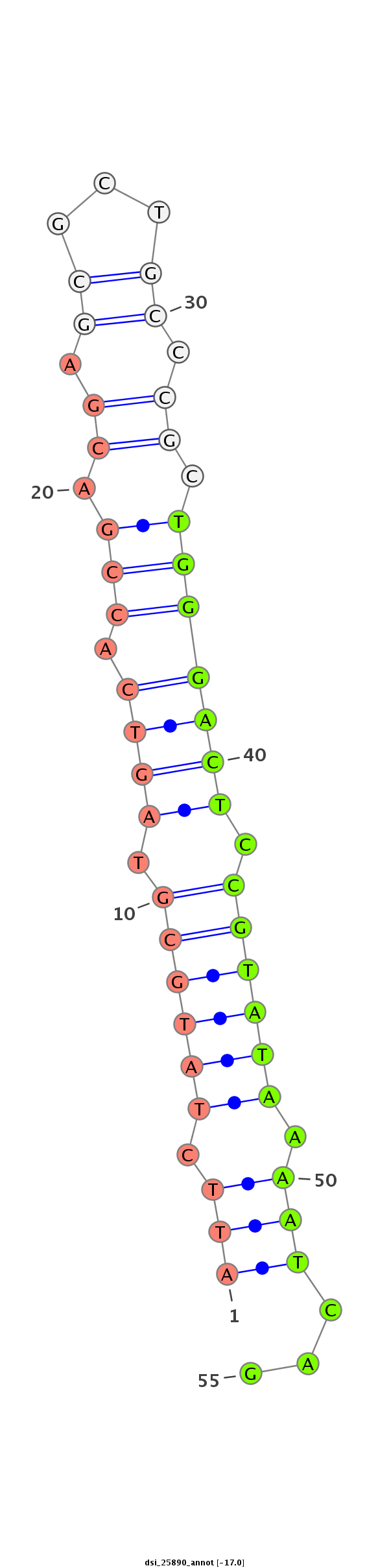
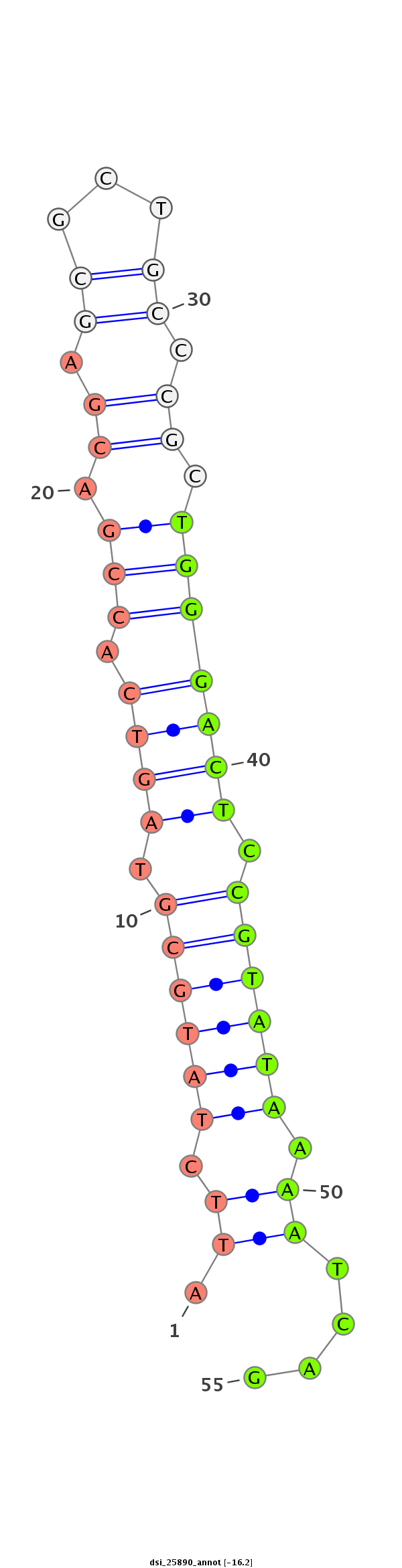
ID:dsi_25890 |
Coordinate:3l:19710268-19710484 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -17.0 | -16.2 |
 |
 |
intergenic
No Repeatable elements found
|
CAACGAAAGTGAACATCACATTTAGCGCTTCGCAGTCGCGGCTTCAACCGGTATGTGTATGTGAATGTGAGTCTGAATCTGAGTCTCAGACAGCCGATATATATGGCCATATATTTTTCTACTTTTGTTCTGTTTCGCCAGTCCCGCACATGCGATGATTGTGGTTTTGTTTTTAGTGGTATTCCATTTTATTCTATGCGTAGTCACCGACGAGCGCTGCCCGCTGGGACTCCGTATAAAATCAGCATAATCAGAAATTCTACCCAGCAGTTCGTTCTAAGCCATCGACGGAGCCGGACGCTCATCGCGCTTTTGGC
**********************************************************************************************************************************************************************************************....((((((.((((.(((.((.((...)).)).))))))).)))))).......************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M024 male body |
O002 Head |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCG..................... | 18 | 0 | 1 | 18.00 | 18 | 17 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCCGGAC.................. | 19 | 0 | 1 | 15.00 | 15 | 9 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TGGGACTCCGTATAAAATCAG........................................................................ | 21 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TGGGACTCCGTATAAAATCAGT....................................................................... | 22 | 1 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGACGCTC.............. | 25 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGACGCT............... | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................TGGGACTCCGTATAAAATCAGC....................................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................ATTCTATGCGTAGTCACCGACGA........................................................................................................ | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................GCTTCGCAGTCGCGGCTT................................................................................................................................................................................................................................................................................. | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGACGC................ | 23 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCCGGACGCTCA............. | 24 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGA................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGACGCTCATC........... | 28 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGG................... | 20 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................GTGAAAGTGAGTCTGAATC.............................................................................................................................................................................................................................................. | 19 | 1 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCCGGACGCTC.............. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCCGGACGC................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCCGGACGCTCAT............ | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................AGCCATCGACGGAGCCGGAC.................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TGTGGTTTTGTTTTTAGTG........................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................CTGGGACTCCGTATAAAATCAGC....................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................CCATCGACGGAGCCGGACGC................ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TAAGCCATCGACGGAGCC...................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGAC.................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGAGCCGGACGCTCA............. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGGATCCGGACGCTCAT............ | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................CCATCGACGGAGCCGGACG................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................AAGAGATGATCGTGGTTTT..................................................................................................................................................... | 19 | 3 | 17 | 0.71 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 0 | 0 | 1 |
| .............................................................................................................................................................ATTGTGGTTTTGTTTTT............................................................................................................................................... | 17 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................TATTATATCCGTAGTCTCCG............................................................................................................ | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................TGAAAGTGAGTCCGAATCTGCG.......................................................................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................GCCATCGACGGAGCTGGA................... | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................GTCCCGCACATACGA.................................................................................................................................................................. | 15 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................CCATATGTTTTTCTGCTTTTGTTA........................................................................................................................................................................................... | 24 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................TTTCGCCACTCCCGCAAA....................................................................................................................................................................... | 18 | 2 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................CAAGAGATGATCGTGGTTT...................................................................................................................................................... | 19 | 3 | 12 | 0.25 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| ............................................................GTGAAAGTGAGTCCGAAT............................................................................................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................AAGCCATCGACGCGGCCG..................... | 18 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................ATACAATCACCATATTCAGA.............................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................AACCGGTCTGTGAATATGAA............................................................................................................................................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................CTAAGCAATCGACGG.......................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................AAGTGAGTCTGAATC.............................................................................................................................................................................................................................................. | 15 | 1 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................CTTCGGAGGCGCGGGTTCA............................................................................................................................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................................................................TGGGAATCCTTATAAACTC.......................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTTGCTTTCACTTGTAGTGTAAATCGCGAAGCGTCAGCGCCGAAGTTGGCCATACACATACACTTACACTCAGACTTAGACTCAGAGTCTGTCGGCTATATATACCGGTATATAAAAAGATGAAAACAAGACAAAGCGGTCAGGGCGTGTACGCTACTAACACCAAAACAAAAATCACCATAAGGTAAAATAAGATACGCATCAGTGGCTGCTCGCGACGGGCGACCCTGAGGCATATTTTAGTCGTATTAGTCTTTAAGATGGGTCGTCAAGCAAGATTCGGTAGCTGCCTCGGCCTGCGAGTAGCGCGAAAACCG
************************************************************************....((((((.((((.(((.((.((...)).)).))))))).)))))).......********************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GTCAGCGCCGAAGTTGGCCATACAC.................................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................AGATCCGAATCAGTGACTG.......................................................................................................... | 19 | 3 | 11 | 0.73 | 8 | 3 | 5 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................CAAGCAAGACTCGGT................................. | 15 | 1 | 15 | 0.47 | 7 | 0 | 0 | 0 | 1 | 5 | 0 | 1 |
| ........................................CGAAGTTGGGCATGCACA................................................................................................................................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................GTCCTATTAGTCCTTAAGA........................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................AGAAAAAGCGGTGAGGGCGTTT....................................................................................................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AGTGGCTGCTGGCGACCGGC.............................................................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................AAAGCGGTGAGGGCGTTT....................................................................................................................................................................... | 18 | 2 | 7 | 0.29 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................TTAGTCGTATTAGTCGTTTCG......................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................AAAGACGAAAACAGGACAAA...................................................................................................................................................................................... | 20 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................AAAGCGGTGAGGGCGT......................................................................................................................................................................... | 16 | 1 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................CAAGCAAGATTCGTT................................. | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................AAGGACGAAAACAAGACAA....................................................................................................................................................................................... | 19 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................GAAGTTGTTCATACACAT.................................................................................................................................................................................................................................................................. | 18 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................CAAGCAAGACTCGGTAC............................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................CGCGTCAGTCGCTGCCCGC..................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................AGACTTCGACTCAGA....................................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:19710218-19710534 - | dsi_25890 | -----CAACGAAAGTGAACAT-------CACATTTAGCGCTTCGCAGTC-G------------CGGCTTCAACCGGTATGTG-----------------------------TATGTG------------AATGTGAGTCTGAATCTGA--------G------TCTC------------------------------------------AGACAGCCGATATA--------TATGGC--------------------CATATATT------------T--TTCTACTTTTGTTCTGTTTCGCCAGTCCCGCACAT------GCG--ATG--AT---TGT--GG--------TTTTGTTTTTAGTGGTATTC--------C--ATTTTATTCTATGCGTAGTCA---CCGACGAGCGCTG--------------------------------------------------------------------------------------------------------------CCCG-------CTG-GGAC------------T-CCGTATAAAATCAGCATAATCAGAAAT-TCTACCCAGCAGTTCGTTCTAAGCCATCGACGGAGCCGGACG---CTCATCGCG-CTTTTG-------GC |
| droSec2 | scaffold_11:30212-30536 - | -----CAACGAAAGTGAACAT-------CACATTTAGCGCTTCGCAGTT-G------------CGGCTTCAACCAGTATGTG-----------------------------AATGTGAGT------------------CTGAATCTGAGTCTCTGAATCTGACTCTC------------------------------------------AGACAGCCGATATA--------TATGGC--------------------CATATATT------------T--TTCTACTTTTGTTCTGTTTCGCCAGTCCCGCACAT------GCG--ATG--AT---TGT--GG--------TTTTGTTTTTAGTGCTATTC--------C--ATTTTATTCTATGCGTAGTCA---CCGACGAGCGCTG--------------------------------------------------------------------------------------------------------------CCCG-------CTG-GGAC------------T-CCGTATAAAATCAGCATAATCAGAAAT-TCTACCCAGCAGTTCGTTCTAAGGCATCGACGGAGCCGGACG---CTCATCGCG-CTTTTG-------GC | |
| dm3 | chr3L:20120431-20120747 - | -----CAACGAAAGTGAACAT-------CACATTTAGCGCTTCGCAGTC-G------------CGGCTTCAACCGGTATATG-----------------------------TATGTG------------AATGTGAGTCTGAATCTCA--------G------TCAC------------------------------------------AGACAGCCGATATA--------TATGGC--------------------CATATATT------------T--TTCTACTTTTGTTCTGTTTCGTCAGTCCCGCACAT------GCG--ATG--AT---TGT--GG--------TTTTGTTTTTAGTGGTATTC--------C--ATTTTATTCTATGCGTAGTCA---CCGACGACCGCTG--------------------------------------------------------------------------------------------------------------CCCG-------CTG-GGAC------------T-ACGTATAAAATCAGCATAATCAGAAAT-TCTACCCAGCAGTTCGTTCTAAGCCATCGACGGAGCCGGACG---CTCATCGCG-CTTTTG-------GC | |
| droEre2 | scaffold_4784:19848625-19848970 - | -----CAACGAAAGTGAACAT-------CACACTTAGCGCTTCGCAGTC-G------------CGGCTTCAACCGGTATAGG-----------TAAG------GGTATGTGTCTGTGAGT------------------CTGAGTCGGG--------G------TCTC------------------------------------------AGACAGCCGATATACAGATATATATGGC--------------------CATATATT------------T--TTCTACTTTTGTTCTGTTTCGCCAGTCCCGCACAT------GCG--ATT--AT---TGT--GG--------TTTTGTTTTTAGAGGTATTCCACTATTCC--ATTTCATTCTATGCGTAGTCA---CCGACGATCGCTG--------------------------------------------------------------------------------------------------------------CCCGCTGCCCGCTG-GGAC------------T-TCGTATAAAATCAGCATAATCAGAAAT-TCTAGGCAGCAGTTCGTTCTAAGCCATCGACGGAGCCGGACG---CTCATCGCG-CCTTTG-------GC | |
| droYak3 | v2_chr3L_random_222:17602-17932 + | -----CAACGAAAGTGAACAT-------CACATTTAGCGCTTCGCAGTC-G------------CGGCTTCAACCGGTATGTG-----------CGTGTATATGTGTATGTGTATGTGAGT------------CTGAGTCTGACTCTGA--------G------TCT----------------------------------------------CAGCCGATATA--------TATGGC--------------------CATATATT------------T--TTCTACTTTTGTTCTGTTTCGCCAGTCCCGCACAT------GCG--ATT--AT---TGT--GG--------TTTTGTTTTTAGTGGTATTC--------C--ATTTTATTCTATGCGTAGTCA---CCGACGACCGCTG--------------------------------------------------------------------------------------------------------------TCCG-------CTG-GGAC------------T-CCGTATAAAATCAGCATAATCAGAAAT-TCTACCCACCAGTTCGTTCTAAGCCATCGACGGAGCTTGACG---CTCATCGCG-CTTTTG-------GC | |
| droEug1 | scf7180000409711:5830324-5830646 + | -----GAACGAAAGTGAACAT-------CACATTTAACGCTTCGCAGTCTG------------CGACTTCAACCGGTATGTG-------------------------------------T------------------CTGAATCCAA--------G------TCTC------------------------------------------TGACAACCGATATATC--TGTATACGAG--------------------TATATAT-------------G--TTGTACTTTTGTTCTGTTTCGCCAGTCTCGCACAT------GCG--ATT--AT---TGTGTGGTTTTG---TGTTTTTTTTAGTTTTATTC--------C--ATTTTATTCTATGCGTAGTCGTAGTCGTCGTCCGCTG--------------------------------------------------------------------------------------------------------------CCCG----CTGCTG-GGAC------------T-CCGTATAAAATCAGCATAATCGGAAAT-TCTACCCAACAGTTGGTTCTAAGCCACCGACGGAGCCGGACG---CTCATCGCGGTTTTTG-------GT | |
| droBia1 | scf7180000302193:2898613-2898898 - | -----GAACGAAAGTGAAGAT-------CACATTTAGCGCTTCGCAGTC-G------------CGGCCTCAACCGGTACGAG-------------------------------------T------------------------CTCA--------G------TCTC------------------------------------------TGACAACCAATATA--------CAT----------------------------------------------TTTTACTTTTGTTCTGTTTCGCCAGTCTCGCACAT------GCG--ATT--AT---CGT--GC--------TTGTGTTTTTAGTTGTATTC--------C--ATTTCATTTTATGCGTAGTC------GTCGTACGCTG--------------------------------------------------------------------------------------------------------------CCCG-------CTG-GGAC--------CCACT-CCGTATAAAATCAGCATAATCGGAAAT-TCTACCCAGCAGTTGGTTCTAAGCCACCGTCGGAGCCGGACG---CCCATCGCG-CTTTCG-------GC | |
| droTak1 | scf7180000415868:70754-71071 - | C----GAACGAAAGTGAAGAT-------CACATTTAACGCTTCGCAGTC-G---CTTCAGT--CGGCTTCAACCGGTATGTG-----------------------------TCTGTGAGT------------------CCGATTCCGA--------A------TCCG------------------------------------------AGACAACCAATATA--------TA------------------------------TT------------T--TCCTACTTTTGTTCTGTTTCGCCAGTCTCGCACAT------GCG--ATT--AT---TGT--GGTTTTTTGTTTTTGTTTTTAGTTGTATTC--------C--ATTTTATTCTATGCGTAGTC------GTCGTGCGCTG--------------------------------------------------------------------------------------------------------------CCCG-------CTG-GGAC--------TCACT-CCGTATAAAATCAGCATAATCAGAAAT-TCTACCCAGCAGTTGGTTCTAAGCCATCGTCGGAGCCGGACG---CTCATCGCG-CTTTTG-------GC | |
| droEle1 | scf7180000491193:4424044-4424219 - | -----CAACGAAAGTGAACAT-------CACATTTATAGCCTCGCAGTC-G------------CGGCTTCAACCGGTATGTG-----------TTTG--------------AGTCTGAGTCCGAGTCTGAGTCTGAGTCTGAGTATGA--------G------TCTC------------------------------------------TGACAGCCCATATA--------TAT-------------------------------------------T--TTGTACTTTTGTTCTGTTTCGCCAGTCTCGCATAT------GCG--ATT--AT---TGT--GG--------CTTTGTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000770922:2686-2971 + | -----GAACGAAAGTGAACAT-------CACATTTATAGCTTCGCAGTC-G------------CGGCTTCAACCGGTATGTG-----------------------------TCTGAG------------ATTTTGAGTCTGAGTCTGG--------T------TCTC------------------------------------------CGTCAGCCCATATA--------TAT-------------------------------------------T--TTGTACTTTTGTTCTGTTTCGCCAGTCGCGCACAT------GCG--ATT--AT---TGT--GG--------CTTTGTTTTTAGTTGTATTC--------C--AT-TTATTCTATGCGTAGTC------GTCGACCGCCG--------------------------------------------------------------------------------------------------------------TCCG-------CCGGAGTC------------C-CAGTATAAAATCAGCATAATCGGAAAT-TCTACCTAGCAGTTGGTTCTAAGCCATCGACGAGGCCGGTCG---------------------------- | |
| droFic1 | scf7180000454113:2189950-2190239 + | -----GAACGAAAGTGAACAT-------CACGTTAAACGCTTCGCAGTC-G------------CGGCTTTAACCGGTATCAG-------------------------------------T----------------------------------------------C------------------------------------------TGACAGACGATATA--------TATG----------------------------TTTTTTTTTCTCATT--TTGTACTTGTGTTCTGTTTCGACAGTCGCAT--AT------GCGAGATT--AT---TGC--GG--------CTTTGTTTTTAGTTGTATTC--------C--ATTTTATTCTATGCGTAGTC------GTCATCCGACG--------------------------------------------------------------------------------------------------------------ATCG-------CTG-GGAC------------T-CCGTATAAAATCAGCATAATCAGAAAT-TCTACCCAGCAGTTGGTTCTAAGCCATTGACGGAGCCGGACG---CTCATCGTG-CTTTCG-------GT | |
| droKik1 | scf7180000302686:570167-570506 - | -----CAATGAAAGTGAACAT-----CGCACATTAAATGCTTCGCAGTC-G------------CCGCT-------GTATGTGCGGTGCCATGCTGTG--------------CGTGTGTGC------------CCGGGTCTGGCTCTGG--------GTCTG--TCTC------------------------------------------TGACAGCCAATATA--------TTT--------------TGTTGCTGGCTTATTTT------------A--TTTCACTTTTGTTCTGTTTCGCGAGTCTCGTATAT------GCG--ATT--AT---TGT--GG--------TTTTGTTTTTAGTTGTATTCCA---TTCC--ATTTTATTCTATGCGCAGCC------GT---CCGCTG--------------------------------------------------------------------------------------------------------------CCCA-------CTG-GGAC------------TCCCGTATAAAATCAGCATAATCAGAAAT-TCTACCAAGCAGTTGGTTCTAAGCCGTTGACGGAGCCGGACGCAGCTCATCGCG-CCTCTG-------GT | |
| droAna3 | scaffold_13337:5024816-5025160 + | -----GAACGAAAGTGAACATCGAATCGCACATTAAACGCTTCGCAGTC-A---CT---GTCGCGGTTTTAACCG--------------------------------------------T------------------CTGA--------------G------CC--GCCCGTTTGAGGTTTTTTTTTTCTCGTTTCATTTTTTTTTACTGACAGCCAATATA--------TTT--------------TGTTGCTGG-----T--------------TTATTTTACTTTTGTTCTGTTTCGCGAGTCTCGT--AT------GCG--ATT--AT---TGT--TG--------TTTTGTTTTTAGTAGTATTC--------C--ATTTTATTCTATGCGTAGT------------CCGCTG--------------------------------------------------------------------------------------------------------------GCCG-------CTG-GGACTTGCGGTCCCCGG-CCATATAAAATCAGCATAATCAGAAAT-TCTACCGAGCAGTTGGTTCTAAGCCATTGTCGGAGCCGGACG---CTCATCGCC-TCTCTG-------GC | |
| droBip1 | scf7180000396371:355273-355626 + | -----GAACGAAAGTGAACATCGAATCGCACATTAAACGCTTCGCAGTC-AACGCT---GTCGCGGTTTTAACCG--------------------------------------------T------------------CTGA--------------G------CC--GCCCGTTTGAGGTTTTT-TTTTCTCGTTTCATTTTTTTTTACTGACAGCCAATATA--------TTT--------------TGTTGCTGG-----T--------------TTATTTTACTTTTGTTCTGTTTCGCGAGTCTCGT--AT------GCG--ATT--AT---TGT--TG--------TTTTGTTTTTAGTAGTATTC--------C--ATTTTATTCTATGCGTAGT------------CCGCTG--------------------------------------------------------------------------------------------------------------GCCG-------CTG-GGACTTGCGGTCCCCGG-CCATATAAAATCAGCATAATCAGTAAT-TCTACCGAGCAGTTGGTTCTAAGCCATTGTCGGAGCCGGACG---CTCATCGCC-TCTCTGTGGCCTGGT | |
| dp5 | XR_group6:4691463-4691701 - | -----TG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT--------------TTACTTTACTTTTGTTCTTTTTCGCGTGTTTCGTA--T------GCG--ATTCGATTGTTGT--TGCTG-----TTTTGTTTTTAGTTGTATTT--------C--ATTTTATTCTATGCGTAGT------------CCGCCG---------------------------------------TCTACCGTCGGCAGACG------------------------------------------------------G---CCGTCCGTTGGGGACTTGTGGGCTCTAT-CCATATAAAATCAACATAATCGGAAAT-TCTACCAAGCAGTTGATTCTAAGCAATTGACGGAGCCGGACG---CTCACTCCG-CCTCTG-------GC | |
| droPer2 | scaffold_9:2992088-2992327 - | -----TG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT--------------TTACTTTGCTTTTGTTCTTTTTCGCGTGTTTCGTA--T------GCG--ATTCGATTGTTGT--TGCTG-----TTTTGTTTTTAGTTGTATTT--------C--ATTTTATTCTATGCGTAGT------------CCGCCG---------------------------------------TCTACCGTCGGCAGACG------------------------------------------------------G---CCGTCCGTTGGGGACTTGTGGGCTCTAT-CCATATAAAATCAACATAATCGGAAATTTCTACCAAGCAGTTGATTCTAAGCAATTGACGGAGCCGGACG---CTCACTCCG-CCTCTG-------GC | |
| droWil2 | scf2_1100000004511:1277612-1277961 - | TGTGTGAGTGAGTGTGAACG--GAGTC------------------AGTC-A---------------------------------------------------------------------------------------------------------ATTT------------------------------------TA---------------------TATATG--TATAAACAGACGTATGTATTATATTGT---CGTA-------------------TTTTA-TTTTGTTCTTTTC-----------------------TG--ATG--GC---TGC--TGC-------TTTTGTTTTTAGTTGTATTT--------C--ATTTTATTCTATGCGTAAA------------TCGTTATTCTGGCGACTGCGCCAATAGCACGCTGCTGCGTCGCCGTC-AGCGTCGGC-GTCGGCGGTGTCTCTGTCTGTGTCTGGGCCTGCTTTGGGGTCTGGCTCTGACTCTGGTTCTGG-CTTTGGGG-GTCT--------TCCAC-ATATATAAAATCAGCATAATCAGTTAT-TCTACCAAGCAGTTGATTCTAAGCAATTGACAAAGCCGCACA---C------------------------ | |
| droVir3 | scaffold_13049:18297140-18297287 - | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTTTTTTAGTTGTTTTC--------C--ATATTATTTTATGCGTACT------------ACGCTG--------------------------------------------------------------------------------------------------------------TCCGG-CTCTGTGG-GCTA------------T-CCATATAAAATCAACATAATCAGAAAT-TCTACCGAGCAGTTGATTCTATGCGGTCGACGAAGCCGGACG---CGCT--GCC-TTCTGG-------GG | |
| droMoj3 | scaffold_6680:19080444-19080653 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGTTCTTT-TTACGTGTTTCGTA--TACAGCTACG--ATC--TT---TAT--ATATGTTTTTCATTATTTTTTGTTGTATTTTA---TTTCGTATTTCGTTCTATGCGTACA------------ACGCTG--------------------------------------------------------------------------------------------------------------TCCGG-CTCCACGG-GCAA------------T-CCATATAAAATCAGCATATTCAGAAAT-TCTACCGGACAGTCGATTCTATGCGGTCAACGGAGTCGGACG---CGCTGCCTT-CTTGGG-------GC | |
| droGri2 | scaffold_15110:9346884-9347060 + | ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGTTCTTT-TTACGTGTTTCGTA--TACTCGTACG--A------------------------TATTTTTTTTAGTTGTTGTC--------C--ATTTTATTCTATGCGTACT------------ACGCTG--------------------------------------------------------------------------------------------------------------TCCG-------GGG-GCTG------------T-CCATATAAAATCAACATAATCGGAAAT-TCTAGCGAGCAGTTGATTCTATGCGATCGACGAAGGCGGACG---CGCTGC----CTTCTG-------GG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 06:54 PM