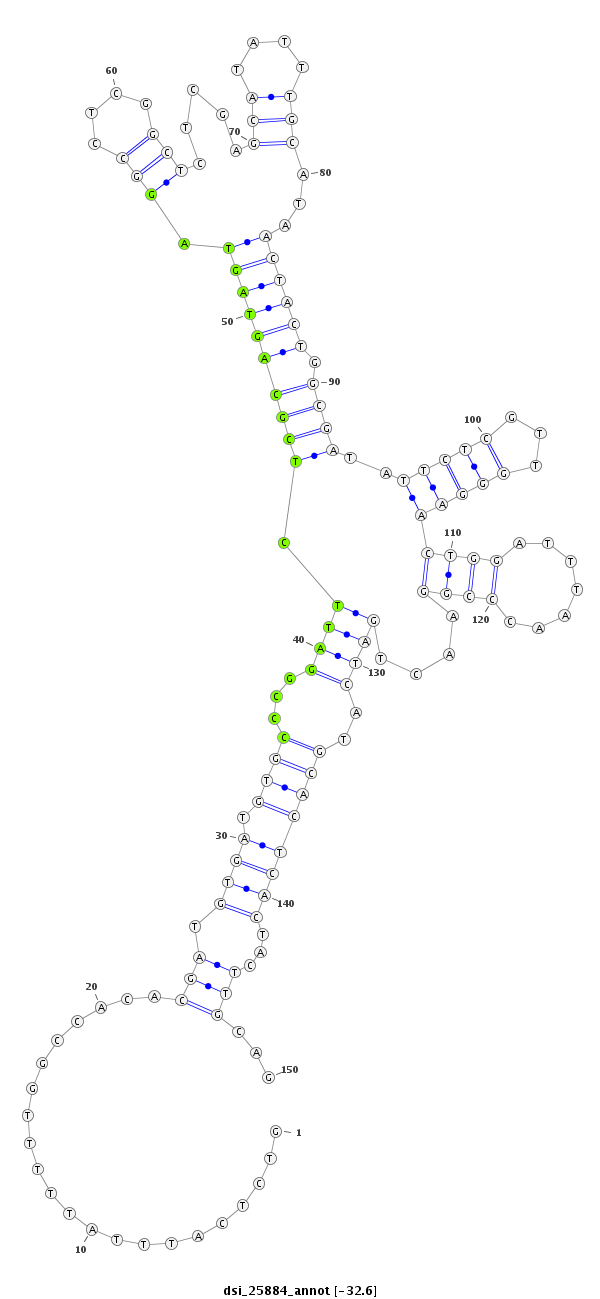



ID:dsi_25884 |
Coordinate:3l:11480218-11480367 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -32.6 | -32.6 | -32.6 |
 |
 |
 |
exon [3l_11479504_11480217_-]; CDS [3l_11479504_11480217_-]; intron [3l_11480218_11482168_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GTGGTGCTAGTCCAACCGATCCACATCGCACCGCTGCGACTTTAGGGGCCGTCTCATTTATTTTTGGCCACACGATGTGATGTGCCCGGATTCTCGCAGTAGTAGGCCTCGGCTCTCGAGCATATTTGCATAACTACTGGCGATATTCTCGTTGGGAACTGGATTTAACCCGGAACTGATCATGCACTCACTACTTGCAGGCAACGACACCGCCTCCGACAGCACTTCCTACTATGCGGAATACGATCCC **************************************************......................(((.((((.((((...((((.((((((((((.(((....))).....(((....)))...)))))).))))..(((((...)))))((((.......))))....))))..))))))))...)))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR902009 testis |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................CGCTGCGACTTTAGGGGC......................................................................................................................................................................................................... | 18 | 0 | 1 | 10.00 | 10 | 9 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................CCCGGATTCTCGCAGTAGTAG................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................CGCTGCGACTTTGGGGGC......................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TACTGGCAACGACACCGCCTCCGACCG............................ | 27 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................ATCGCACCGCTGCGACTTTAGGGGC......................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................TGAATTTAACCCGGA............................................................................ | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................TGTGATGTGCTCGGA................................................................................................................................................................ | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CACCACGATCAGGTTGGCTAGGTGTAGCGTGGCGACGCTGAAATCCCCGGCAGAGTAAATAAAAACCGGTGTGCTACACTACACGGGCCTAAGAGCGTCATCATCCGGAGCCGAGAGCTCGTATAAACGTATTGATGACCGCTATAAGAGCAACCCTTGACCTAAATTGGGCCTTGACTAGTACGTGAGTGATGAACGTCCGTTGCTGTGGCGGAGGCTGTCGTGAAGGATGATACGCCTTATGCTAGGG
**************************************************......................(((.((((.((((...((((.((((((((((.(((....))).....(((....)))...)))))).))))..(((((...)))))((((.......))))....))))..))))))))...)))...************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TATTGGGCCTTGACTAGTACG................................................................. | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................AATCCCCGGCTGAGT.................................................................................................................................................................................................. | 15 | 1 | 3 | 1.33 | 4 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| ...................................................................................................................................................................................................................TGGAGGCTGTCGTGAAGGAT................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................ATATTGGGCCTTTACTAGTAGGT................................................................ | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................ACGGGCCTAAGAGTGTCAT..................................................................................................................................................... | 19 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................ATCCCCGGCTGAGTGA................................................................................................................................................................................................ | 16 | 2 | 12 | 0.17 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .............TTGGCAAGGTGTAGGCTGGC......................................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................TTGGGCCTTGGCTAG..................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................ACGGACCTATGAGCGGCAT..................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............TCGCTAGATGTAGTGTGGC......................................................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................GGTGTCCTACACTAC........................................................................................................................................................................ | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................CTGAAATCGACGGCGGAGTA................................................................................................................................................................................................. | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................AGTGGGTCTTGACTAGAAC.................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................CTGACCAGTACGAGAGTG........................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:14 AM