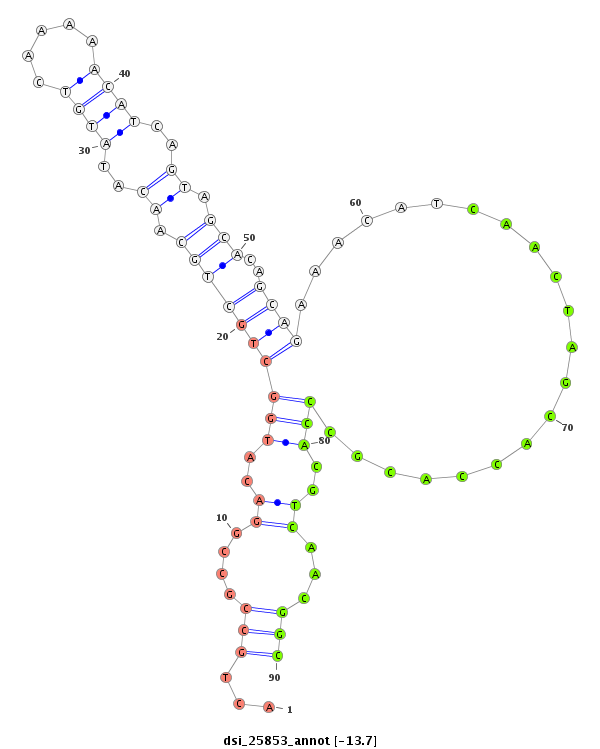
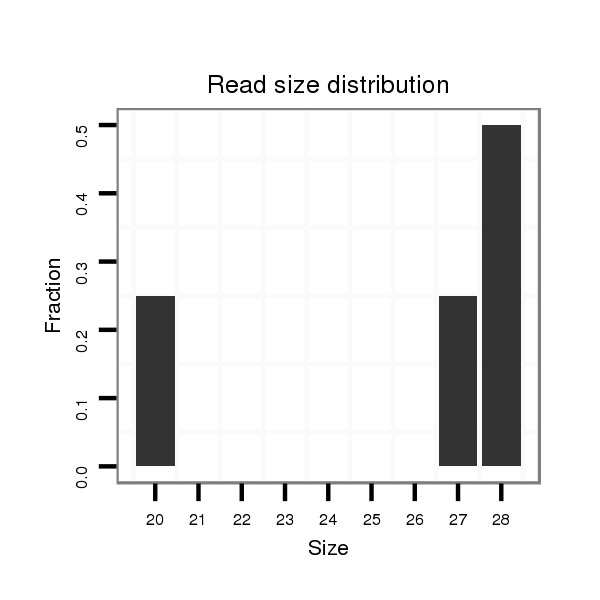
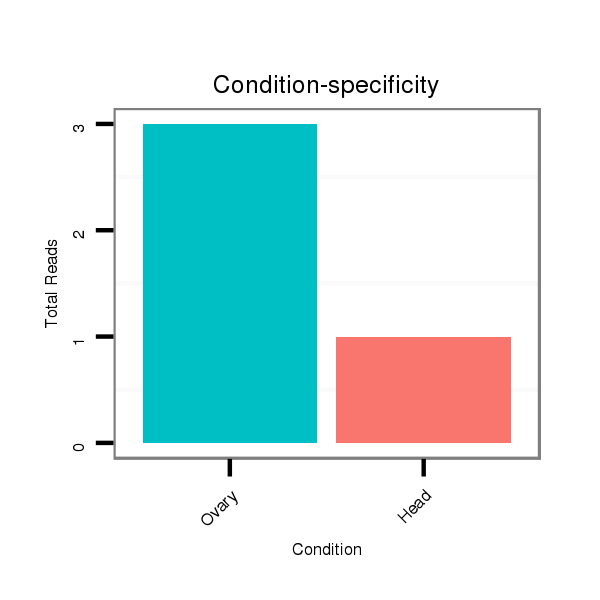
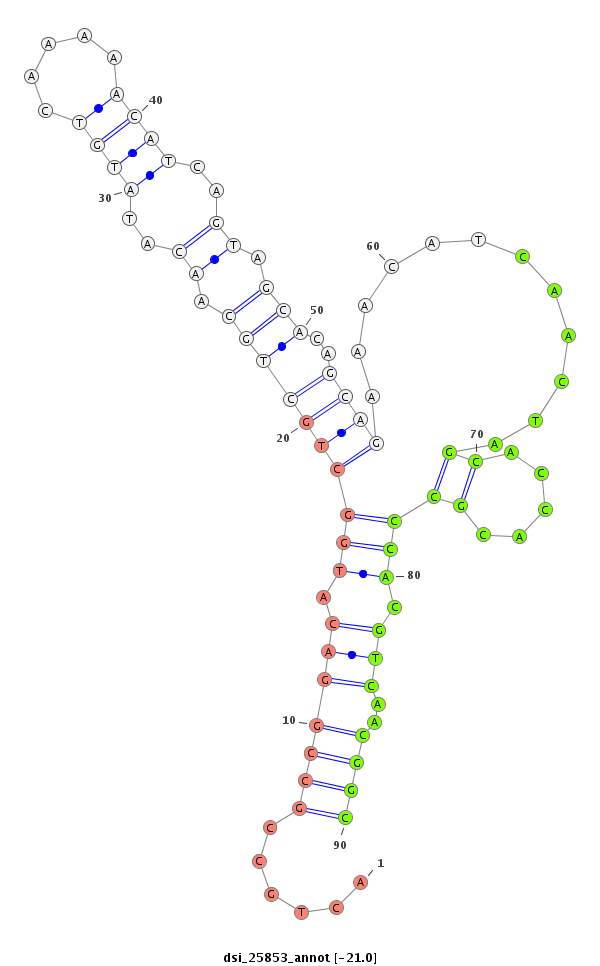
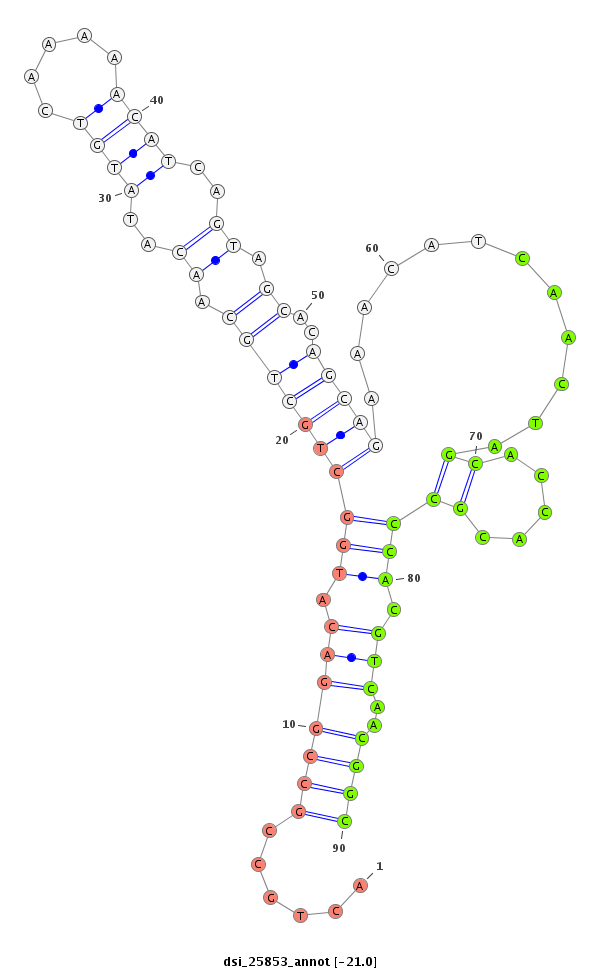
| ............................................................................................................................CTCGGGCTGCTCGATGGTC.......................................................................................................................................................................... |
19 |
3 |
19 |
9.47 |
180 |
127 |
0 |
46 |
0 |
3 |
0 |
0 |
0 |
3 |
0 |
1 |
| ....................................................CTCGGGCTGCTCGATGGTC.................................................................................................................................................................................................................................................. |
19 |
3 |
19 |
9.47 |
180 |
127 |
0 |
46 |
0 |
3 |
0 |
0 |
0 |
3 |
0 |
1 |
| ....................................................CTCGGGCTGCTCGATGGT................................................................................................................................................................................................................................................... |
18 |
2 |
2 |
2.50 |
5 |
1 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| ............................................................................................................................CTCGGGCTGCTCGATGGT........................................................................................................................................................................... |
18 |
2 |
2 |
2.50 |
5 |
1 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................TCGGGCTGCTCGATGGT................................................................................................................................................................................................................................................... |
17 |
2 |
12 |
1.67 |
20 |
0 |
1 |
0 |
1 |
8 |
10 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................TCGGGCTGCTCGATGGT........................................................................................................................................................................... |
17 |
2 |
12 |
1.67 |
20 |
0 |
1 |
0 |
1 |
8 |
10 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGATGGTC.......................................................................................................................................................................... |
20 |
3 |
4 |
1.25 |
5 |
3 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGATGGTC.................................................................................................................................................................................................................................................. |
20 |
3 |
4 |
1.25 |
5 |
3 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGATGGT................................................................................................................................................................................................................................................... |
19 |
2 |
2 |
1.00 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................TAGTACGGCAGGACTG............................................................................................................................................................................................................. |
16 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGATGGT........................................................................................................................................................................... |
19 |
2 |
2 |
1.00 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGACGGTC.................................................................................................................................................................................................................................................. |
20 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGACGGTC.......................................................................................................................................................................... |
20 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................................GCCGCTGACAACGGGA...................................................................... |
16 |
0 |
2 |
0.50 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGACGG.................................................................................................................................................................................................................................................... |
18 |
1 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................................................................................................................................................................................................................................................................ACCACCGGGGGTCACT. |
16 |
1 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGACGG............................................................................................................................................................................ |
18 |
1 |
3 |
0.33 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................CTCGGGCTGCTCGATGG............................................................................................................................................................................ |
17 |
2 |
11 |
0.27 |
3 |
0 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................CTCGGGCTGCTCGATGG.................................................................................................................................................................................................................................................... |
17 |
2 |
11 |
0.27 |
3 |
0 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGATGG.................................................................................................................................................................................................................................................... |
18 |
2 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGATGG............................................................................................................................................................................ |
18 |
2 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................TAGTATAGAGTTTTAGTAGTC............................................................................................................................... |
21 |
3 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................TCGGGCTGCTCGACGG............................................................................................................................................................................ |
16 |
1 |
11 |
0.18 |
2 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................TCGGGCTGCTCGACGG.................................................................................................................................................................................................................................................... |
16 |
1 |
11 |
0.18 |
2 |
0 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGATG..................................................................................................................................................................................................................................................... |
17 |
2 |
12 |
0.17 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................GGCGATGCCCCTGCCCACT................................................................................................................................................................................................................................................................... |
19 |
3 |
6 |
0.17 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGATG............................................................................................................................................................................. |
17 |
2 |
12 |
0.17 |
2 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................GTCTACAGTTTTTGAAGTTAT............................................................................................................................. |
21 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ..................................................TACTCGGGCTGCTCGACGG.................................................................................................................................................................................................................................................... |
19 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................TACTCGGGCTGCTCGACGG............................................................................................................................................................................ |
19 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..............................................................................................................................CGGGCTGCTCGATGGT........................................................................................................................................................................... |
16 |
2 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................CGGGCTGCTCGATGGT................................................................................................................................................................................................................................................... |
16 |
2 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................CCTATACCGGCGACGTTG................................................................................................................................................. |
18 |
2 |
7 |
0.14 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTTCTCGATGGTC.................................................................................................................................................................................................................................................. |
20 |
3 |
7 |
0.14 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTTCTCGATGGTC.......................................................................................................................................................................... |
20 |
3 |
7 |
0.14 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................................GGATGGCAAAGAAGG.......................................................... |
15 |
1 |
15 |
0.13 |
2 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......................................................CGGGCTGCTCGACGGT................................................................................................................................................................................................................................................... |
16 |
1 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................CGGGCTGCTCGACGGT........................................................................................................................................................................... |
16 |
1 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGAAGGTC.................................................................................................................................................................................................................................................. |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGAAGGTC.......................................................................................................................................................................... |
20 |
3 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................................................................AGGGCAAACAAGGAA........................................................ |
15 |
1 |
10 |
0.10 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................TGATGGTGGTCCGGGTGC.......................................................................................... |
18 |
2 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................ACGCGGGCTGCTCGACGG.................................................................................................................................................................................................................................................... |
18 |
2 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACGCGGGCTGCTCGACGG............................................................................................................................................................................ |
18 |
2 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....GTGACTGTTAGTTATTAAAAC............................................................................................................................................................................................................................................................................................... |
21 |
3 |
13 |
0.08 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..........................................................................................GGTAGTCTGACAGGACT.............................................................................................................................................................................................................. |
17 |
2 |
14 |
0.07 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................TAGTCATCGTGTCGGGT................................................................................................................... |
17 |
2 |
17 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................................................................GGATGGCAAAGAAGGGA........................................................ |
17 |
2 |
19 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................TCGGGCTGCTCGACG............................................................................................................................................................................. |
15 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................ACTCGGGCTGCTCGAT.............................................................................................................................................................................. |
16 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................GGCTGCTCGATGGTGACGA...................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................ACTCGGGCTGCTCGAT...................................................................................................................................................................................................................................................... |
16 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................GTGCCCGCTCGGGCTG.................................................................................................................................................................................... |
16 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................GGCTGCTCGATGGTGACGA.............................................................................................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .............................................GTGCCCGCTCGGGCTG............................................................................................................................................................................................................................................................ |
16 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................TCGGGCTGCTCGACG..................................................................................................................................................................................................................................................... |
15 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |