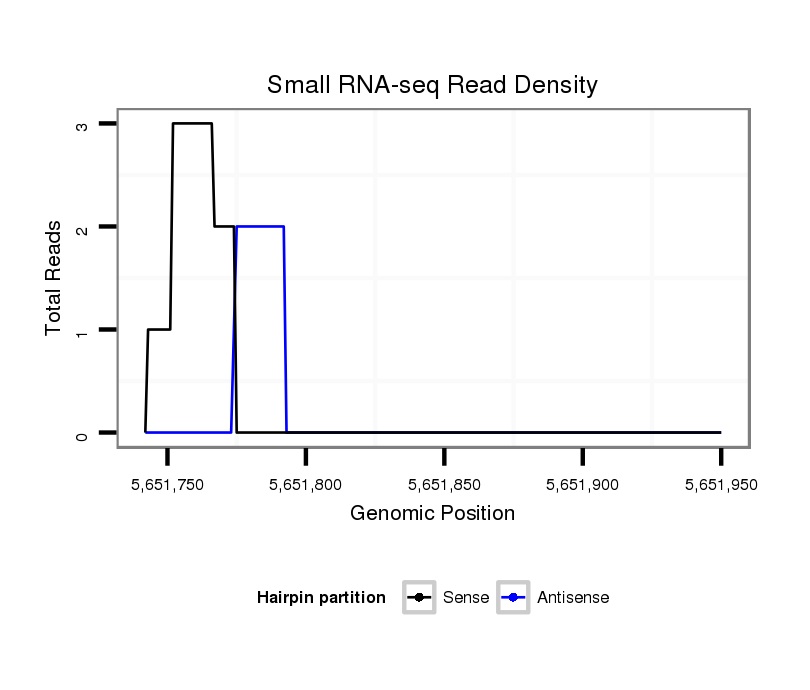
ID:dsi_25623 |
Coordinate:2r:5651792-5651900 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5651674_5651791_+]; CDS [2r_5651674_5651791_+]; CDS [2r_5651901_5652109_+]; exon [2r_5651901_5652109_+]; intron [2r_5651792_5651900_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------################################################## TTCACGTCCGTGGAGAAGAACGCCGATGGAACCATCATCTTCAGCTACAGGTGAGTGCCGGGAGCCTGAGGAGCAGGCCTTTGATTAGGTTAGCACTTGCCCCTCAGAGCAGCCAGTGTGAATATGTCTGATACGATCGATCCCTATCGAGCGATGCAGATGTCTGCACGAGTCTCAAATCTTTCCGCCGGGCCGCTCCATCTGGTGCA **************************************************......((.((..((((((((.((((((((......)))......))))).)))))).))..)).))..........(((...((.(((((....))))).))...)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M025 embryo |
M053 female body |
SRR902008 ovaries |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................ATCGACCGATCCTGATGTCT............................................ | 20 | 3 | 4 | 10.00 | 40 | 19 | 16 | 1 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................ATCGACCGATCCTGATGTC............................................. | 19 | 3 | 9 | 4.22 | 38 | 13 | 17 | 3 | 3 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................GAGGAGCCTGCGTTTGATTAG......................................................................................................................... | 21 | 3 | 2 | 3.00 | 6 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGGAGCCTGCGTTTGATTAG......................................................................................................................... | 20 | 3 | 4 | 2.25 | 9 | 5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........TGGAGAAGAACGCCGATGGAACC................................................................................................................................................................................ | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .TCACGTCCGTGGAGAAGAACGCCG........................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TCGACCGATCCTGATGTCT............................................ | 19 | 3 | 16 | 0.56 | 9 | 5 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........GGTGGATAAGAACGCGGATGGA................................................................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TATCGACCGATCCTGATGTC............................................. | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................CGGATGGAACCATCATATTCT...................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................GGAGCCTGCGTTTGATTAG......................................................................................................................... | 19 | 3 | 14 | 0.29 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGAGGGCCGAGAGGCTGA............................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............AGAAGAACCCCCATGGA................................................................................................................................................................................... | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................AATCGACCGATGCTGATGTC............................................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TGAGACGTTCGGTCCCTA............................................................... | 18 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................GCTTCCCGCCGGGCCGC............. | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................AACCCTCATCTTCAGTT................................................................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................GTTCGATCCCTTTCGCGC......................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...AGGTCCGAGGAGAAGATCG........................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
AAGTGCAGGCACCTCTTCTTGCGGCTACCTTGGTAGTAGAAGTCGATGTCCACTCACGGCCCTCGGACTCCTCGTCCGGAAACTAATCCAATCGTGAACGGGGAGTCTCGTCGGTCACACTTATACAGACTATGCTAGCTAGGGATAGCTCGCTACGTCTACAGACGTGCTCAGAGTTTAGAAAGGCGGCCCGGCGAGGTAGACCACGT
**************************************************......((.((..((((((((.((((((((......)))......))))).)))))).))..)).))..........(((...((.(((((....))))).))...)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|
| ................................GTAGTAGAAGTCGATGTCC.............................................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| .................................TAGTAGAAGTCGATGTCC.............................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 |
| ..............................TAGTAGTGGAAGTCGATGTCCT............................................................................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| ...............................AGTAGTAGAAGTCGATGTCCTG............................................................................................................................................................ | 22 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .................................................................................................................................CTTTGCTAGCTAGGG................................................................. | 15 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 |
Generated: 04/23/2015 at 08:52 PM