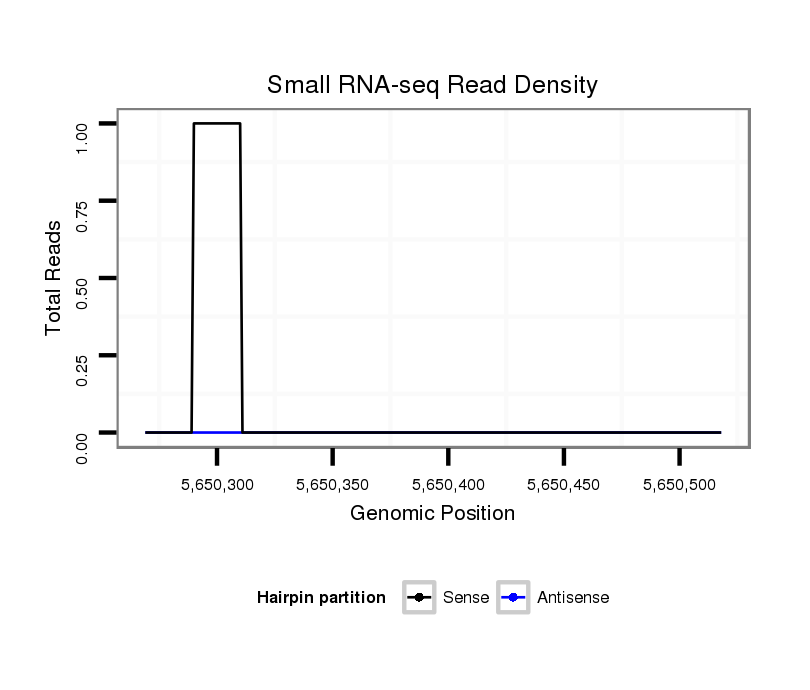
ID:dsi_25506 |
Coordinate:2r:5650319-5650468 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5650004_5650318_+]; CDS [2r_5650004_5650318_+]; intron [2r_5650319_5651673_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TCCCCAGCTGCCCATCTACATTGCTCAGCCTAGCGCGAAAAAGCCAGAAAGTAAGTTCCGATTGCCTAACGAGAGTGGAGAGCACACTGAGGACAATGCACCCCGAATTATCTAAACAAAAAGAATGTGATAGATTTACAGACTTGTTCCGGTATTGACTTGCAAACATTTTACTTGACTTCGATTGTCCAGGAAAGTGAGCCAGAGTAAAGGTCATTTTACTTCCCATTTTTAAGGTCAAATCCATTAT **************************************************((((((..((((.(((.((((((((((.......))))..((........))........(((........))).(((((.....)))))..))))))..)))))))))))))...(((((..((.(((.......))).))..))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
M023 head |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .....................TGCTCAGCCTAGCGCGAAAAA................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................ACCTGTTCAGGTATTGAC........................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................CCAGGAAAGGGAGCCA.............................................. | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................AATTGTTCAGGTATTGAC........................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................TCTACTTGTCTTCGATCGTC............................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 |
|
AGGGGTCGACGGGTAGATGTAACGAGTCGGATCGCGCTTTTTCGGTCTTTCATTCAAGGCTAACGGATTGCTCTCACCTCTCGTGTGACTCCTGTTACGTGGGGCTTAATAGATTTGTTTTTCTTACACTATCTAAATGTCTGAACAAGGCCATAACTGAACGTTTGTAAAATGAACTGAAGCTAACAGGTCCTTTCACTCGGTCTCATTTCCAGTAAAATGAAGGGTAAAAATTCCAGTTTAGGTAATA
**************************************************((((((..((((.(((.((((((((((.......))))..((........))........(((........))).(((((.....)))))..))))))..)))))))))))))...(((((..((.(((.......))).))..))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
M024 male body |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................TCTAAATGTCAGAACACG..................................................................................................... | 18 | 2 | 7 | 1.14 | 8 | 0 | 5 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............TAGATGGAACGAGTCGGAA.......................................................................................................................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCTAAATGTCAGAACA....................................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CTAAATGTCAGAACACGGA................................................................................................... | 19 | 3 | 10 | 0.50 | 5 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........CGGGTAGATGTAGAGAGACG............................................................................................................................................................................................................................. | 20 | 3 | 10 | 0.50 | 5 | 0 | 3 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................CTAAATGTCAGAACACGG.................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................ACCTCTCGTGTTACCCCTCTT.......................................................................................................................................................... | 21 | 3 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCTAAATGTCAGAACATG..................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TCTAAATGTCAGAAC........................................................................................................ | 15 | 1 | 12 | 0.17 | 2 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................CTAAATGTCAGAACACG..................................................................................................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................ACAGAGTGCTGTCACCTCT......................................................................................................................................................................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................AGGGTAGAAATTCCA............ | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............TAGATGTCAGGAGTCG............................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 08:49 PM