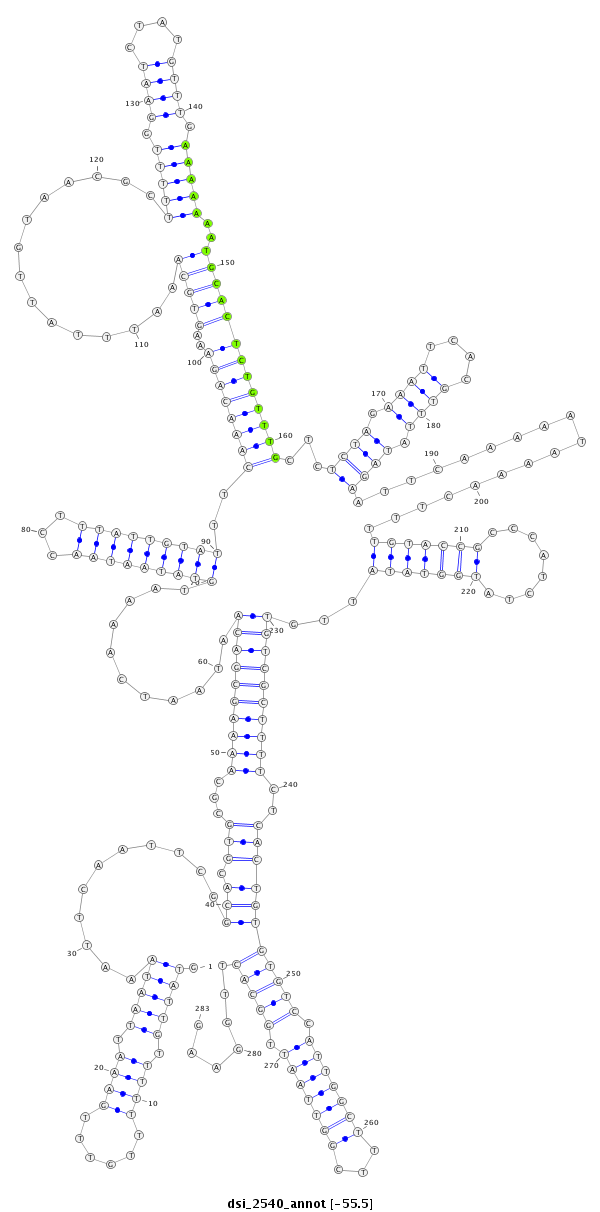
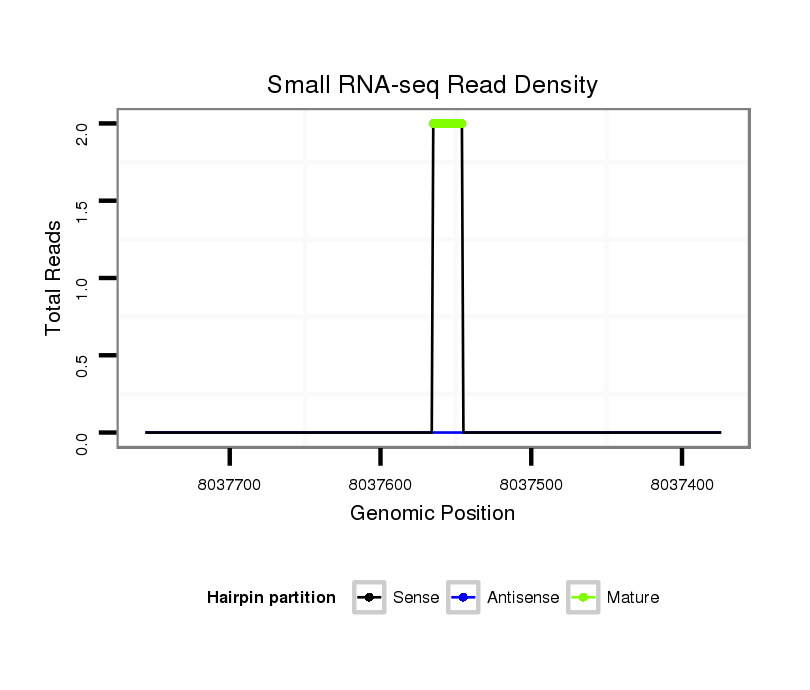
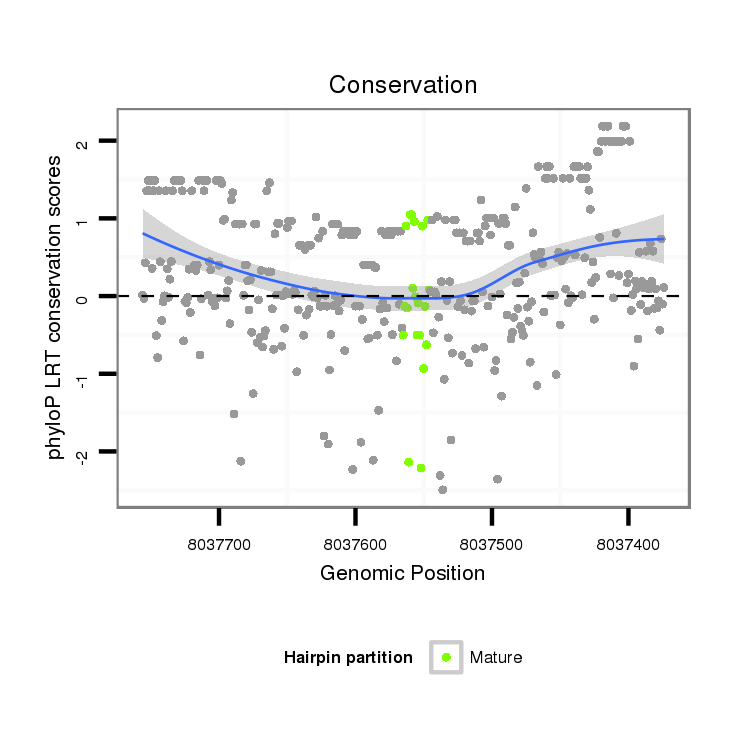
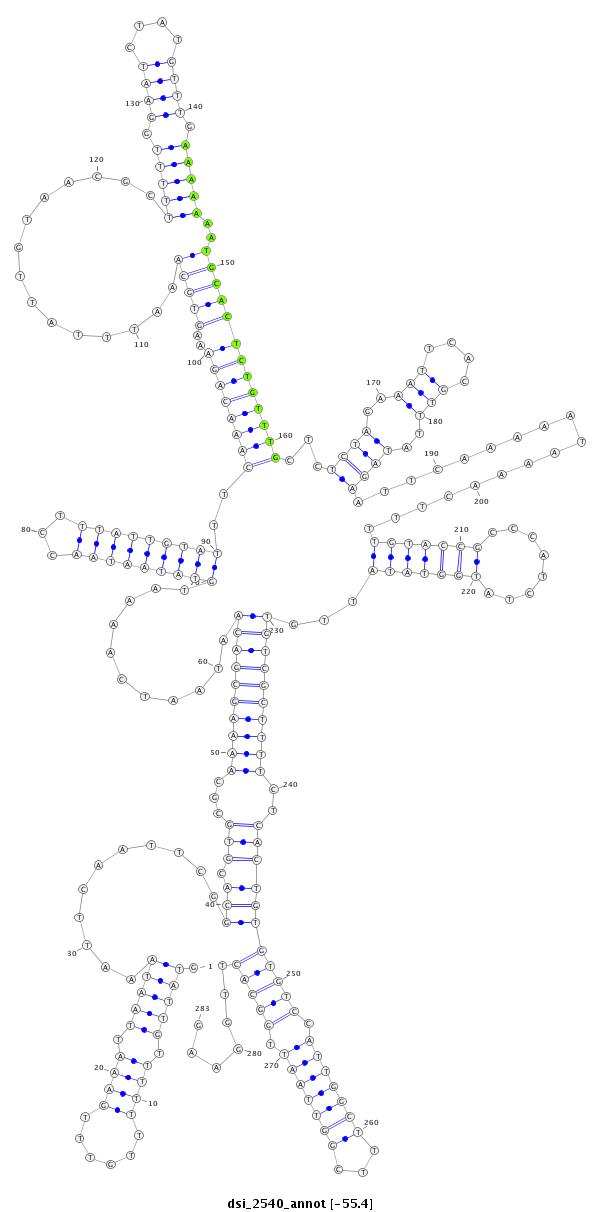
| droSim2 |
x:8037374-8037756 - |
dsi_2540 |
A-AAGTCAAGAAGCAA----------CCG--AGCAGATAATACAG------CTCGGG-------A------ATTAAAC---AAAAGT----AT-TGTTT---------------TTTTGTTTGAAATTAATAAAT--TC-----AATTCGG---------------------------CA----CGTG--CGC------------------------------AAAAGCGACAATAAT--------------------------------------------------------------------------------CAAAATGTATA----------ATAACCTTTAT-----------TGTATTTCAAACAGAAAGTGCAAATTTATTGTAACGCTTTTTGGAA-------------------TCTATGT-TTG----AA--------------------------------------------------------------AA-----------------------------AAATGC---------------------------------------ACTCTGTTTGCTCTCTAGAAATTCACGTT----------------------------------------TATAGAATTCAA-AAATAAA----------------------------------------------------------------------ACTTTTGTACCGCCCAT--CTAT------GGTATATTG------------------TGT--------CGCTTTTCTCACTGTGTGTCC-----------ATTGGCTTTCGGT------TAATTGGCACTTGG-AAGTTGGAAGTTGCCCGGCTGGTTTCTGCTTTC---------------CGATC----TG-----------GGATCGCCACTTG-------- |
| droSec2 |
scaffold_35:315078-315469 - |
|
A-AAGTCAAGAAGCAT----------CCG--AGCTGATAATACAG------CTCGGG-------A------ATTAAAC---AAAAGC----GTATTTT-----------------TTCGTTTGAAATTAATAAAT--TC-----AATTCGG---------------------------CA----CGTG--CGC------------------------------AAAAGCGACAATAAT--------------------------------------------------------------------------------CAAAATGTATAATGTACGCGAATAACCTTTAT-----------TGTATTTCAAACAGAAAGTGCAAATATATTGTAACGCTTTTAGGAA-------------------TCTATGT-TTG----AA--------------------------------------------------------------AA-----------------------------AAATGC---------------------------------------ACTCTGTTTGCTCTCTACACATTCACGTT----------------------------------------TATAGAATTCAA-AAATAAA----------------------------------------------------------------------ACTTTTGTACTGCCCAT--CTAT------GGTATATTG------------------TGT--------CGCTTTTTTCACTGTGTGTCC-----------GTTGGCTTTCGGT------TAATTGGCACTTGG-AAGTTGGAAGTTGCCCGGCTGGTTTCTGCTTTC---------------CGATC----TG-----------GGATCGCCACTTG-------- |
| dm3 |
chrX:8466990-8467382 - |
|
A-AAGTCAAGAAGCGA----------CCG--AGCAGATAATACAG------CTCGGG-------A------ATTAAAC---AAATGC----GTATTTT----------------TTTTGTTTGAAATCAATAAAT--CC-----AAGTTGG---------------------------CA----C--------------------------------------------GACAATAAT--------------------------------------------------------------------------------CAAAATGTATAATGAACGCGAATAACCTTTAT-----------TGTATTTCAAACAGAAAGTACAAATTTATTTTAATGCTTTTAGGAA-------------------TCTATAT-GTGCAAAAA--------------------------------------------------------------AA-----------------------------AAATGC---------------------------------------ACTATGTTTGCTCT--ACACTTTCAAGTT----------------------------------------TATAGAATTCAAAAAATAAA-------------------------------------------------------------------TGCACTTTTGTACTCCCCAT--CTATATTGTATGTATATTG------------------TGT--------CGCTTTTCTCACTGTGTGTCC-----------GTTGGCTTTCGGT------TAATTGGCACTTGG-ATGTTGGAAGTTGCCCGGCTGGTTTCTGCTTTC---------------CGATC----TG-----------GGATCGCCACTTG-------- |
| droEre2 |
scaffold_4690:6645196-6645578 + |
|
A-GTGTCATGAAGCAT----------CTG--AACAGATAATACGG------CTCGGG-------A------ATTGAAC---AACAGC----GTTTGTTTT---------------------TAAAATTAATAAGT--CG-----AATGTGG---------------------------CA----CGTG--CGC------------------------------AAAAGCGACACTATT--------------------------------------------------------------------------------AAAAAAGTATA-TGTGCTCGAATCACCATTGTTTTTTTTATTTTTTATTTCAAACAAAAAGTTCAATTGAATTGCAAAGCAGGTAAGAA-------------------TCCATGT-TTA----TA--------------------------------------------------------------AG-----------------------------GAATGT---------------------------------------ACTATTAATGCTCC--AAGTTTTTGTGTT----------------------------------------TCTAGAA------AAATA---------------------------------------------------------------------------TATGTATTTCTGAT--CTAT------GGCATATTG------------------AGT--------CGG--TTTTCTCAGTGTGTCC-----------GTTGGCTTTCGTTTCAATTTAATTGGCACTT--------GGAAGTTGCCCGGCTGGTTTCTGCTTTC---------------CGATC----TG-----------GGATCTCCAATCG-------- |
| droYak3 |
X:12844790-12845164 + |
|
A-GTGTCATGAAGCAT----------CCG--AACAGATAATACGT------CTCGGA-------A------ATTGAAC---AAAAGC----GTTTGTTTT---------------------TCAAATAAGTAATC--CG-----AATGTGG---------------------------CA----CGTG--CGC------------------------------AAAAGCGACACTAAT--------------------------------------------------------------------------------CAAAAAGTATA----------------TTTCA-----------TAAATTGCAAACAGAAAGTACAAATTAATTGTAATGCACTTAAGCA-------------------TCCATGT-TTA----TA--------------------------------------------------------------AG-----------------------------CAATGC---------------------------------------ACTGTGAATGCTCT--ACATTTTTACATT----------------------------------------TCTAGAA------AAGTA---------------------------------------------------------------------------TATGTATTTCTGATCTCTAT------GGAAGTCGG------------------AGT--------CGG--TTTTCTCAGTGTGTCC-----------ATTGGCTTTCGGT------TAATTGGCACTTGA-AAGTTGGAAGTTGTCCGGCTGGTTTCTGCTCTCTGCCTGGTCTCTGTCAGATC----TG-----------GGATCGCCGATCG-------- |
| droEug1 |
scf7180000408744:85492-85936 - |
|
A-ATGTCAAGACCCAT----------CTG--GACAGATAATACAG------CTTGGG-------A------TTTAAAC---AAAAGCCATCGTTTGTTTTTT-----TTTATTATTATTTTTT---------AATTTCGGATGT-----GTC-------------------CAGAGTAAGAGCGCGTG--TC-TGATCTCAA---------ATTTGCAAATAATAATTTAGCACTGATCG--------------------------------------------------------AAAATGTCACACCT------------------------------------------------------------------------------------------------------------------------------------------------TATAA-TTGAAAATATTAAACCGTATCGCAGTAATTACT---ATAT------------------------------------AAATG--------TGCCTTGTATTAAATCACTACCAGTGAACA-----------GC------------------TTCTACTACAATTACGTTGAGTTCAATTCCCTTAAAAGTTTATTTA-TTTAACAAATAAA-------------------------------------------------------------------TGTTATTTT-AACAACTAAT--CGATTTTGGAATTAT-TTA------------------AAT--------AGG--TTCTCTCAGTGTGTC------------GTTGGCTTTCGGC------TAATTGGCACTTGG-AAGTTGGAAGTTGCCCGGCTGGTTTCTGCTTTC---------------CCATC----TG-----------GGATCGCCAATCG-------- |
| droBia1 |
scf7180000302069:959993-960088 + |
|
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTCTCAGTGTGCCG-----------CCTCGCTTGCGAC------TAATTGCCTCTTGG-AAGTTGGAAGTTGCCCGGCTGGTTTCTGCTCTC---------------AGGTC----TG-----------GGAGCGCCAATCG-------- |
| droTak1 |
scf7180000414549:186515-186616 - |
|
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCTCTCAGTGTCTCT------------CTGGCTTTCGAC------TAATTGCCTCTTGG-AAGTTGGAAGTTGCCCGGCTGGTTTCTGCTCTC---------------CGATC----TG-----------CGATCGCCAATCCAATACAA- |
| droEle1 |
scf7180000491006:1916350-1916438 - |
|
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTTCAGTGTGTCT-----------GTTGGCTGTTGGC------TAATTGACACTT--------GGAAGTTGCCCGGCTGGTTTCTGCTCAG---------------CGATC----TG-----------CGATCTGGGATCG-------- |
| droRho1 |
scf7180000779514:121180-121445 + |
|
A-GTGTCAAGACCCATCTCATCTCATCTG--GACAGATAATACTG------CTCCGG-------A------ATTGAAC---AAAAGCCAGCGATTGTTTATATTGT------------TATTA---------AATTTTCGAGGG-----GCA-------------------CAGAGAAAGAGCGCGTG--CCCCGTACAGAAACAAACACAATTTCAAACTGATAATCCAACACTGAAA--------------------------------------------------------------------------AATTATTAAATGGACA----------AAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTAATGTTTTTCAT---TTTAAGC----TAATGC---------------------------------------CTGCTTTTTAATATTTAAAAATACATATT----------------------------------------TATACATTTAAG-TTTTAAA----------------------------------------------------------------------A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droFic1 |
scf7180000451789:267282-267744 - |
|
A-GTGTCAAGAACCAA----------ATG--GACAGATAATACAG------ACCGGG-------A-------TTGCAC---AAGAGCAAGCGATTGTTTTTATTAT------------TGTTA---------AATTTTGGAGGG-----GCA-------------------GAAAGAAGAAGCGCGTG--CC-TGTACACAAACAAATACGATTTTAAATTGATAATCCGACACTGATATAAAAATAAGTGTTTAATCTTTTAAAATTACTTTTTTAATATCCAATAACAATTTTAT----TTAATTCATAATAAA-A----------------------------------------------------------------------------------------------------------------------------------------TATAACTTGGAA-------------------------------------TT---------------TTTTATTTTAT---------------------------------------------------------------------------------TGC------------------------------------------------------ATCTTTTACACGATCTGTCAAGATTGTTAAAAATGAAAACCAAACCATTTAGAAGAATTCCAAAATTGT-------------------------------AATATATAGTTTCATAAAAAATCGCCTAATAATGAATACAA--TTTTCTCAGTGTTTCT-----------GTCTGCTCTTGGC------TAATTGGCACTTGG-CTGTTGGAAGTTGCCTGTCTGGTTTCTGCTCTC---------------GGATC----TG-----------GGATCGCTAATCG-------- |
| droKik1 |
scf7180000302344:1705961-1706369 - |
|
G-GTGTCAAGACGCAT----------TTGCTGGGAGATAATACATCGTCGGCTCAGG-------G------AAT-AAC---AAAAGCCATCGATTGCTTCTATTATAATT-CG-------------------TATTTTTGAGAG-----ACCAGTTTTTGAAAGGTTTTTTAAGAAAAAAAATACATATATTCTATTAAG------------------------------------------------------------------TTACTTT--------ACAAAAATAATTTTAAAAAAGTAACACATAAAAAATA----------------------------------------------------------------------------------------------------------------------------------------TATAA-TTGGAAAT------CTGTA---GAGTAATTGTTCAAATATTAA--AAATTAAT------------CTTAGACTTCGGAAAGCCTTTGTATGCCTTA-----A----------------AGCTTACTTTT------CTCAACA--CA---------------------------CAATTCCCT---------------------------------------------------------------------------------------------------------------------------------------T------------------AAT--------TAA--ATCTCTCAGTGTGACTGG---------CTTGGCTTTTGGT------TAATTGGCACTTGGGAAGTTGGAAGTTGCCCGGCTGGTTTCTGCGATC---------------TGTGATCTCGC-----------GCATCGCCGATTG-------- |
| droAna3 |
scaffold_13334:933354-933735 - |
|
GAGTGTCAAGAACCAT----------CTG--GAGAGATAATACGG---CAGACCAGGGGCAGGGACTTCAAATTCGACACCAAAAATTTATATTTGTTTTGA-----ATATTTTTTTTTTTTTAAATTAA---------GAGGGAAGGCGG---------------------------GTTGCGCGTG--TA-TGCACCGTGAAAAATT-----------------------------------------------------------------------------------------------------------------------------------TAAT-----T-----------AAAGTTTTACTCAAAAAGTTCCAAGTA-------------------CTTCTTTCCAA------------------------TTAGTTAGATATTTTAGTTTGGCT-------------------------------------TT---------------TT---TTTTAA---------------------------------------------------------------------------------CGA------------------------------------------------------ATCTTTTT---------------------------------------------------------TTGT-------------------------------AATAT-TT-TTTCAGACCTTATCGGATCAT--------CATATTGTTCTACGTGTGACTGA----CGATTGTTTACATTTGGT------TGATTGGCACT-------TTGGAAGTTGCCAGGCGGGTTTACGAGATA---------------CGATC----GCATATGT-TTGAAGCCCTCCACTTG-------- |
| droBip1 |
scf7180000396733:577449-577852 - |
|
A-GTGTCAAGAATCAT----------CTG--GAGAGATAATACGG---CAGACCAGGGGCAGGGACTTCGAATTCGACACCAAAAA---TTATTTGTTTTTA-----ATT-TGTTTTTTTTTTAAATCAA---------GAGGGAAGGCGG---------------------------GTTGCGCGTG--TC-TGCACAGTAAAAAA------------------------------------------------------------------------------------------------------------------------------------TTAATCTGTTT-----------TGTTTATCAA---------------------------TTTTAAGAACTTATATTCAAAGAATGCTTCTATTTTTTG----AAAAGTTTGATCTTT-------------------------------------------------------------TTG--------------------------------------------------------------------GTGCTTTCTTAAAGTAAACATTTTCGA------------------------------------------------------ATCTTTTT---------------------------------------------------------TCGTACATTTTTGTAGACCTTAT--C--------------------------------AGATTAT--------CGTATTGTTCTCTGTGTGACTGACTGACGATTGTTTACTTTCGGT------TGATTGGCACT-------TTGGAAGTTGCCAGGCGGGTTTCTGTGATA---------------CGATC----GCATATGAATTGAAGCCCTCCAGTTA-------- |
| dp5 |
XL_group1e:10802603-10802658 - |
|
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGAAGTTGTCCGGCAGGTTTGCGGCTGC---------------CGACA---CTC-----------GAATAGATATCCG-ATCCGAT |
| droPer2 |
scaffold_14:247286-247334 - |
|
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGAAGTTGTCCGGCAGGTTTGCGGCTGC---------------CGTCA---CTC-----------GAATAGATATCCG-------- |
| droGri2 |
scaffold_14853:6977968-6977996 + |
|
G--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTTTGAAGTTGCCTGGCTGGTTTCTG--------------------------------------------------------------- |