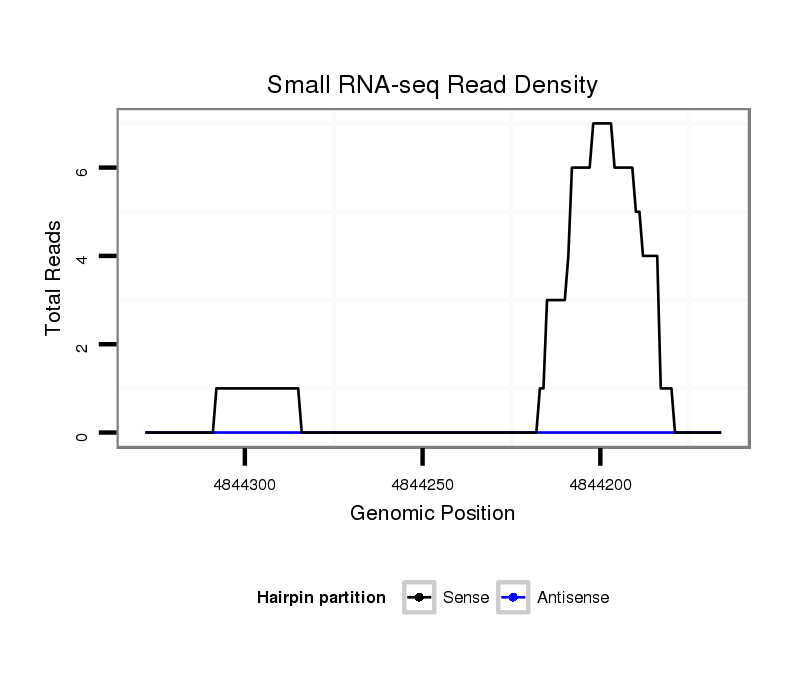
ID:dsi_25221 |
Coordinate:2r:4844216-4844278 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_4844004_4844215_-]; CDS [2r_4844004_4844215_-]; CDS [2r_4844279_4844474_-]; exon [2r_4844279_4844474_-]; intron [2r_4844216_4844278_-]
No Repeatable elements found
| ##################################################---------------------------------------------------------------################################################## CAACCACTTGAGCAACACTCATGGTATGCCTTTGGGCAAGGATGAAAAAGGTACATCAAGGCTATCCCTAAATGTTAATGAACAACCCATTTAAAGATTGTACTGATTTTCAGATGGAGATCAGGCAGCTGTGGCAACGCAAACCTCGCGGCCTACCTCGCCC **************************************************....((((..((.(((..((((((.............))))))..))).))..))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
M023 head |
M024 male body |
M053 female body |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................TCAGGCAGCTGTGGCAACGCAAACC.................. | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................AAAGATGGAGATCAGGCAGC.................................. | 20 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AAGATGGAGATCAGGCAGCTGTGGCCA.......................... | 27 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................ATGGAGATCAGGCAGCTGTGGCAAC......................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................AAGATGGAGATCAGGCAGCTGTGGCA........................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............AACACTCATGGTATGCCTTTGGGCAAGGT......................................................................................................................... | 29 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................AGCTGTGGCAACGCAAACCTCGC.............. | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................ATGGTATGCCTTTGGGCAAGGATG....................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AAAAGATGGAGATCAGGCAGCTGT............................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GGTGATCAGGCAGCTGTGGCA........................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................AGATGGAGATCAGGCAGCTGT............................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ATCAGGCAGCTGTGGCAACGCAAACC.................. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATGGAGATCAGGCAGCTGTGGCAACGC....................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................GGTAGGCCTTTGGGCGAG........................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TAAAGGCTATCCCTAAGGG......................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................CATTGTATGATTTTGGGCAA............................................................................................................................ | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................ATTTAAAGATTGAACAG.......................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTTGGTGAACTCGTTGTGAGTACCATACGGAAACCCGTTCCTACTTTTTCCATGTAGTTCCGATAGGGATTTACAATTACTTGTTGGGTAAATTTCTAACATGACTAAAAGTCTACCTCTAGTCCGTCGACACCGTTGCGTTTGGAGCGCCGGATGGAGCGGG
**************************************************....((((..((.(((..((((((.............))))))..))).))..))))......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TGGGTAAATTTCAAAC............................................................... | 16 | 1 | 13 | 2.92 | 38 | 27 | 1 | 4 | 4 | 2 |
| ..........................................................................................................................................CGTTTTGAGCTCAGGATGGA..... | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................TGGGTAAATTTCAAA................................................................ | 15 | 1 | 20 | 0.35 | 7 | 2 | 0 | 0 | 1 | 4 |
| .....................................................................................GGGTAAATTTCAAAC............................................................... | 15 | 1 | 20 | 0.30 | 6 | 0 | 1 | 5 | 0 | 0 |
| ..TGGTCAACTCGATGTGAGGA............................................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................TGGTTGGGTAAATTTC................................................................... | 16 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................GCGTTTGCAGCGCCG........... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...GGTTAACTCGATGTGAGGA............................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..............................................................ATAGGGTTATACAGTTACTT................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/23/2015 at 07:21 AM