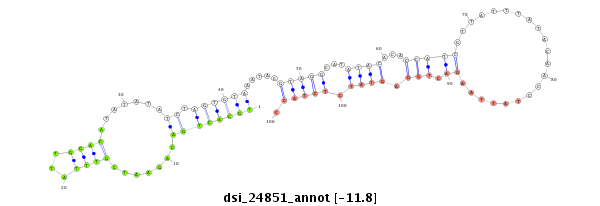
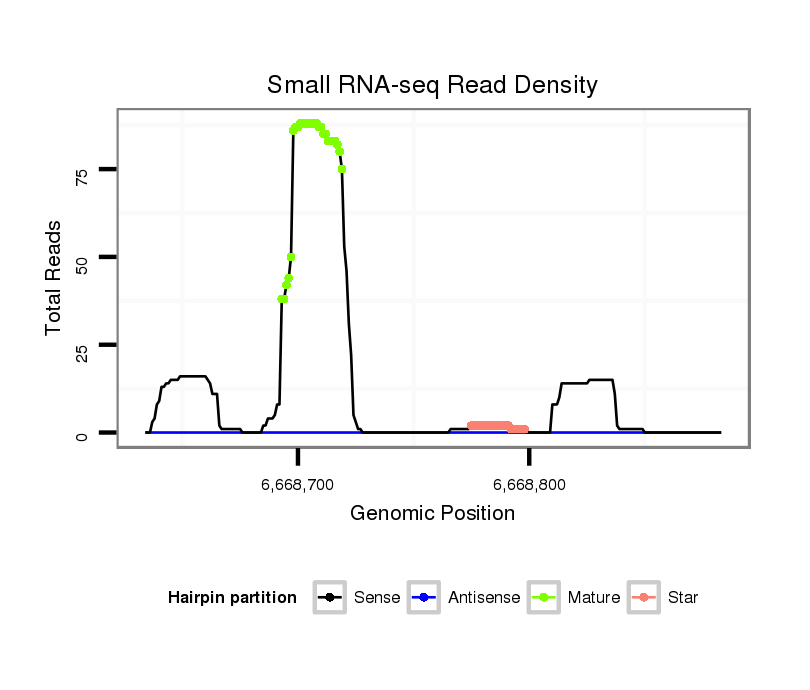
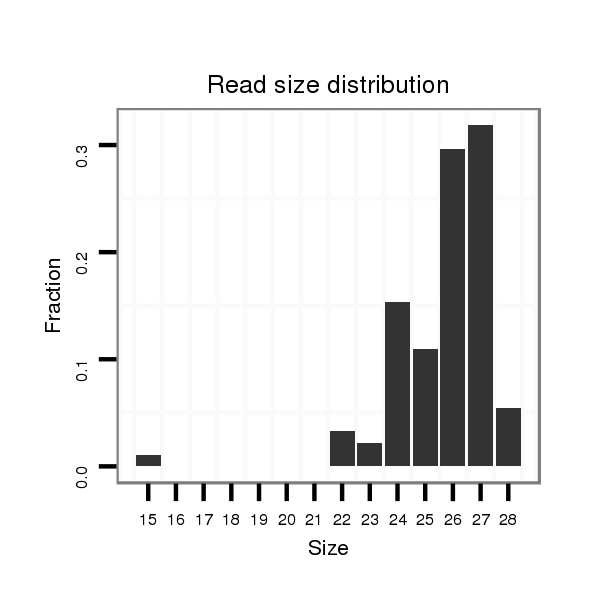
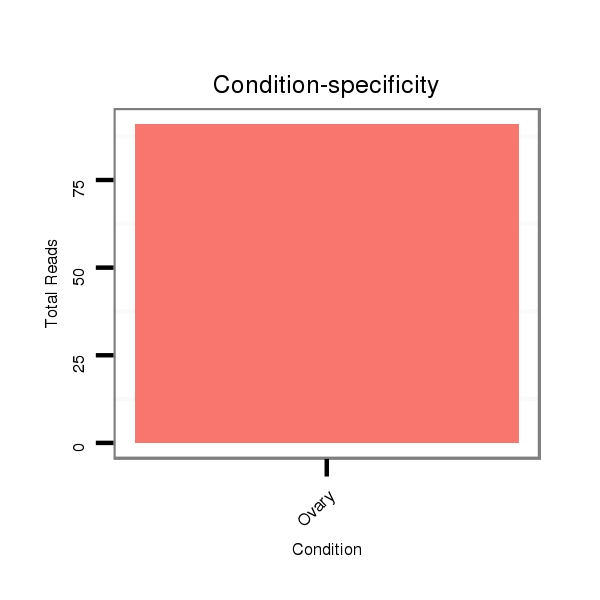
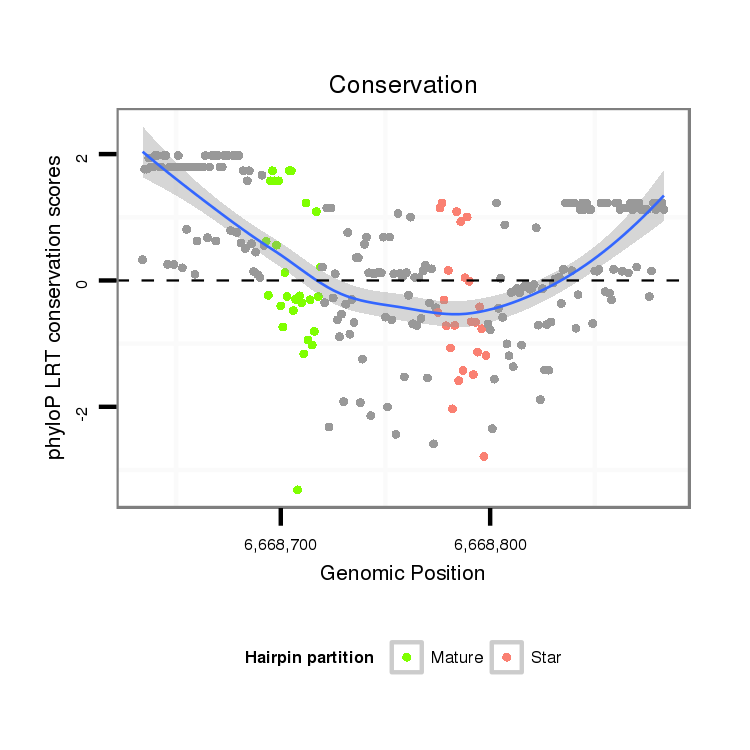
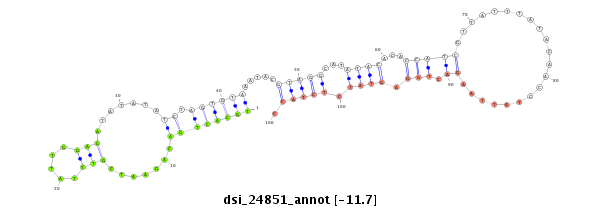
| ...........................................................TGCACTGACAGAATCGTTTATTGGACA.................................................................................................................................................................... |
27 |
0 |
1 |
21.00 |
21 |
21 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATATA................................................................................................................................................................ |
26 |
0 |
1 |
15.00 |
15 |
15 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATA.................................................................................................................................................................. |
24 |
0 |
1 |
10.00 |
10 |
10 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATATAA............................................................................................................................................................... |
27 |
1 |
1 |
8.00 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATAT................................................................................................................................................................. |
25 |
0 |
1 |
7.00 |
7 |
7 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................TACTTCCGAAAGTAGATATGCAATCATT.............................................. |
28 |
0 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......TCGATAGCTGACGGAGCCGGGCCTC.......................................................................................................................................................................................................................... |
25 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................TGCACTGACAGAATCGTTTATTGGAC..................................................................................................................................................................... |
26 |
0 |
1 |
4.00 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...TCCATCGATAGCTGACGGAGCCGGGC............................................................................................................................................................................................................................. |
26 |
0 |
1 |
3.00 |
3 |
2 |
0 |
1 |
0 |
0 |
0 |
0 |
| ...........................................................TGCACTGACAGAATCGTTTATTGGACAT................................................................................................................................................................... |
28 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CACTGACAGAATCGTTTATTGGACATA.................................................................................................................................................................. |
27 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATAA................................................................................................................................................................. |
25 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................TACTTCCGAAAGTAGATATGCAATCAT............................................... |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CTGACAGAATCGTTTATTGGACATATA................................................................................................................................................................ |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................TCCGAAAGTAGATATGCAATCATT.............................................. |
24 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....CATCGATAGCTGACGGAGCCGGGCCTC.......................................................................................................................................................................................................................... |
27 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CTGACAGAATCGTTTATTGGACATAT................................................................................................................................................................. |
26 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................TAAGAATATGCACTGACAGAATCGTT............................................................................................................................................................................. |
26 |
0 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....CATCGATAGCTGACGGAGCCGGG.............................................................................................................................................................................................................................. |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATATATA.............................................................................................................................................................. |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................GACAGAATCGTTTATTGGACATATATA.............................................................................................................................................................. |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........TAGCTGACGGAGCCGGGCCTT.......................................................................................................................................................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................TCCGAAAGTAGATATGCAATCATTT............................................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .......................................................................................................TACGTACGCATATACACACCATCGT.......................................................................................................................... |
25 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................TATGCAATCATTTTTCTCAGTGCA.................................. |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CTGACAGAATCGTTTATTGGACATAA................................................................................................................................................................. |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................CACTGACAGAATCGTTTATTGGACAT................................................................................................................................................................... |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................TTCCGAAAGTAGATATGCAATCAT............................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................TGCACTGACAGAATCGTTTATTGGA...................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....CATCGATAGCTGACGGAGCCGGGCCTCC......................................................................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................AGAATATGCACTGACAGAATCG............................................................................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACAT................................................................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................ACTGACAGAATCGTTTATTGGACATA.................................................................................................................................................................. |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................TATGCACTGACAGAATCGTTTATTGGA...................................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................TATGCACTGACAGAATCGTTTATTGGAC..................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................AGAATCGTTTATTGGACATATATATC............................................................................................................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................AGAATATGCACTGAC...................................................................................................................................................................................... |
15 |
0 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................................................................................................................................................TTCCGAAAGTAGATATGCAATCATT.............................................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................TGCACTGACAGAATCGTTTATTGG....................................................................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATATAAA.............................................................................................................................................................. |
28 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATAAA................................................................................................................................................................ |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................TCCGAAAGTAGATATGCAATCAT............................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................CAGAATCGTTTATTGGACATATAT............................................................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................TATTGGACATATATATCTAGTGTATA................................................................................................................................................... |
26 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................TATTAAGACTGGAGTATCTCTACC..................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ......ATCGATAGCTGACGGAGCCGG............................................................................................................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........GATAGCTGACGGAGCCGGGCCTC.......................................................................................................................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................TATACGTACGCATATACACACCATCGT.......................................................................................................................... |
27 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....CCATCGATAGCTGACGGAGCCGGGCCTC.......................................................................................................................................................................................................................... |
28 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................TATACAACCTATTAAGACTGGAGTAT............................................................................................ |
26 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........TAGCTGACGGAGCCGGGCCTC.......................................................................................................................................................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACATATAT............................................................................................................................................................... |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................TATGCACTGACAGAATCGTTTA........................................................................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CTGACAGAATCGTTTATTGGACAT................................................................................................................................................................... |
24 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............TGACGGAGCCGGGCCTCCATCTGCATA................................................................................................................................................................................................................ |
27 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................ACTGACAGAATCGTTTATTGGACAT................................................................................................................................................................... |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ........................................................ATATGCACTGACAGAATCGTTTA........................................................................................................................................................................... |
23 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................TGACAGAATCGTTTATTGGACA.................................................................................................................................................................... |
22 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................CGTACGCATATACACACCATCGTTATATA.................................................................................................................... |
29 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................CTGACAGAATCGTTTATTGGACATA.................................................................................................................................................................. |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................................................................................................................TAACTTACTTCCGAACGTCGGT......................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................TAGGAGTAACCCCGATGATC. |
20 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ............................................................................................................................................................................................TAGATATGGAATCATT.............................................. |
16 |
1 |
3 |
0.33 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..........................................................................................................................................................................................................................CATACGTTATGTATGT................ |
16 |
1 |
10 |
0.10 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................................CCTATTAAGGCTGGAGGTT............................................................................................ |
19 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .................ACGGAGCTGGGACTCCACCT..................................................................................................................................................................................................................... |
20 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
| ..............................................AAAGGTAAGACTACGGACT......................................................................................................................................................................................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................GTGTACATACGAAGGCA......................................................................................................................................... |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |