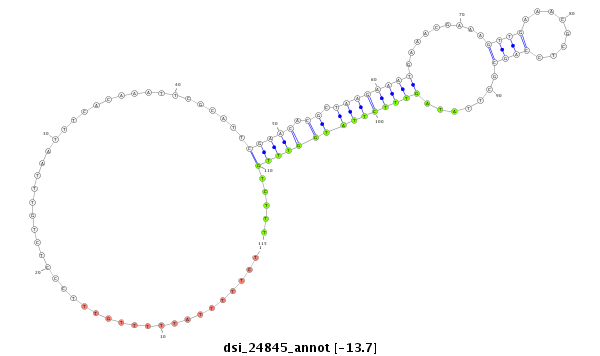
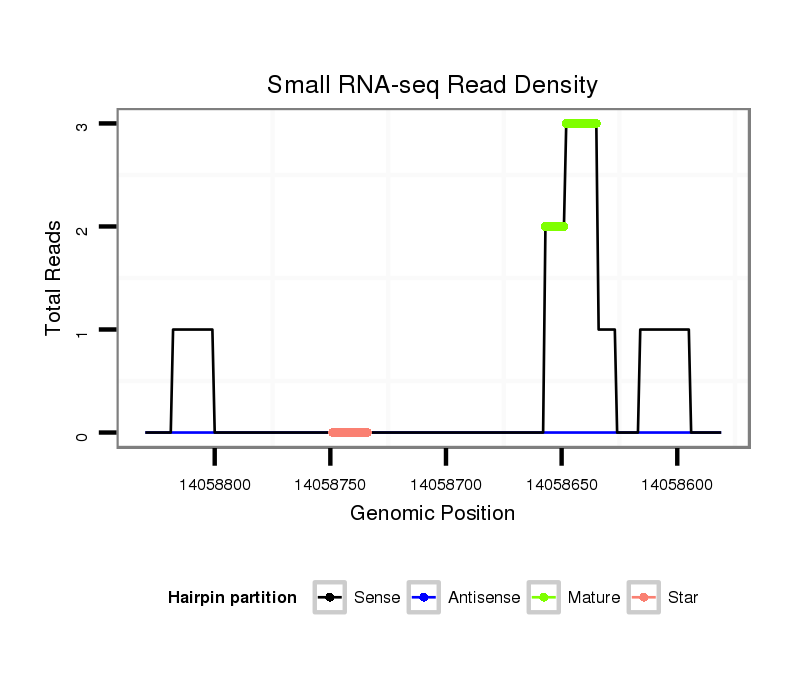
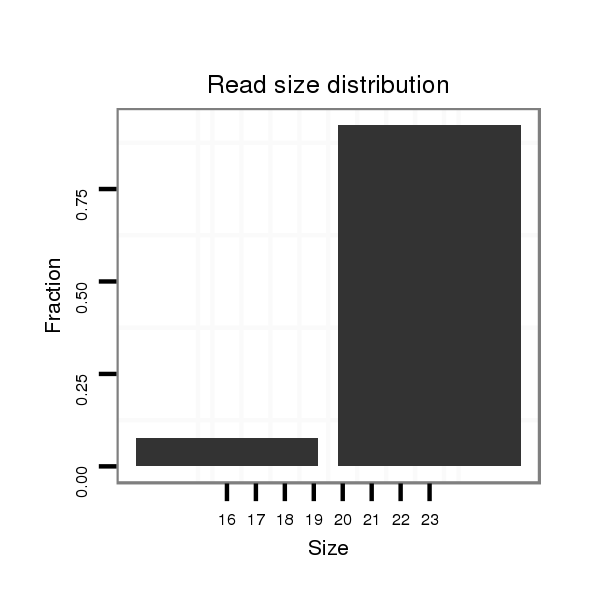
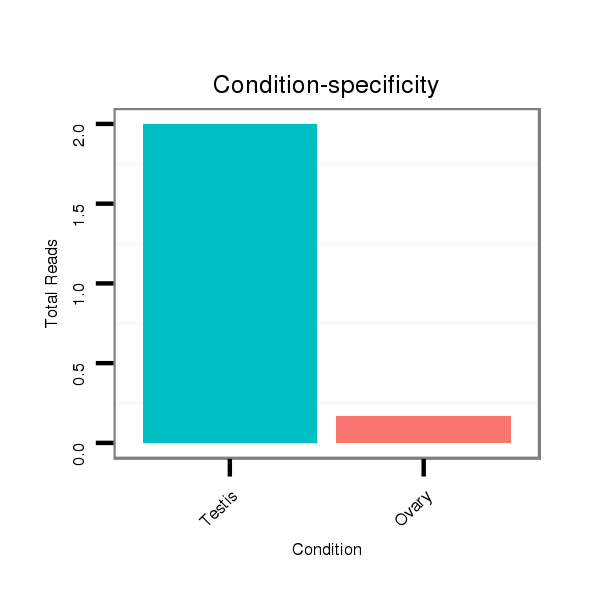
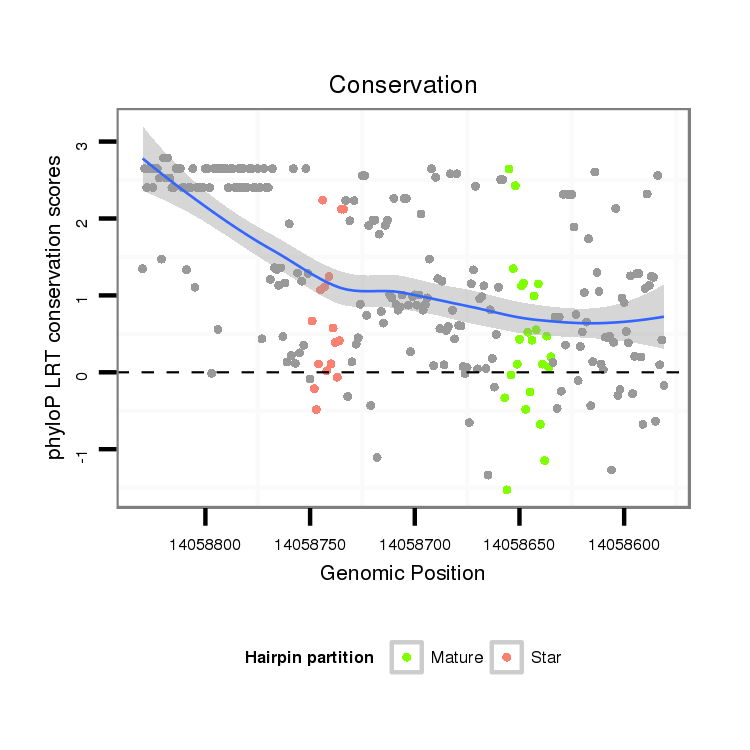
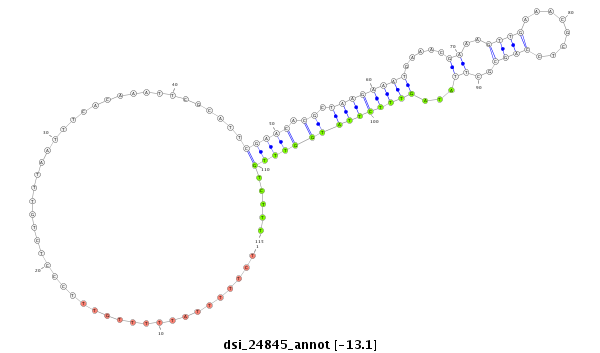
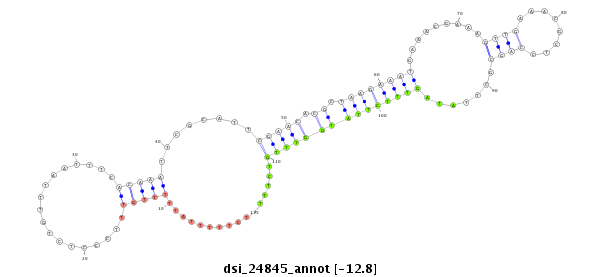
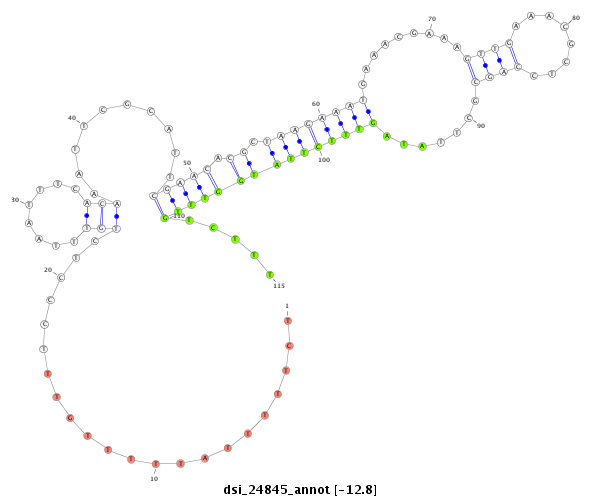
| .................................................................................................................................................................CGAGGTGGCGAATCTCAA....................................................................... |
18 |
2 |
4 |
20.75 |
83 |
83 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................GAGGTGGCGAATCTCAAA...................................................................... |
18 |
2 |
1 |
14.00 |
14 |
14 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................GAGGTGGCGAATCTCAA....................................................................... |
17 |
2 |
8 |
12.25 |
98 |
98 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TGCTAGGTGGCGAATCTCAA....................................................................... |
20 |
3 |
3 |
11.67 |
35 |
34 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................TTGCTAGGTGGCGAATCTCAA....................................................................... |
21 |
3 |
2 |
11.00 |
22 |
14 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CGAGGTGGCGAATCTCAAA...................................................................... |
19 |
2 |
1 |
8.00 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCGAGGTGGCGAATCTCAA....................................................................... |
19 |
2 |
1 |
8.00 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CTAGGTGGCGAATATCAA....................................................................... |
18 |
2 |
4 |
2.75 |
11 |
10 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................TTGAGAGGTGGCGAATCTCAA....................................................................... |
21 |
3 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TCCGAGGTGGCGAATCTCAA....................................................................... |
20 |
3 |
5 |
1.60 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TGAGAGGTGGCGAATCTCAA....................................................................... |
20 |
3 |
2 |
1.50 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................GGTCGCGAATCTCAA....................................................................... |
15 |
1 |
9 |
1.00 |
9 |
8 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TGCTAGGTGGCGAATCTCAAA...................................................................... |
21 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCTAGGTGGCGAATATCAA....................................................................... |
19 |
2 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................TTGGGAGGTGGCGAATCTCAA....................................................................... |
21 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCGAGGTGGCGAATCTCAAA...................................................................... |
20 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TTCGAGGTGGCGAATCTCAAA...................................................................... |
21 |
3 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCAAGGTGGCGAATCTCAA....................................................................... |
19 |
3 |
11 |
0.73 |
8 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................AGGTCGCGAATCTCAA....................................................................... |
16 |
1 |
3 |
0.67 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................ACTAGGTGGCGAATATCAA....................................................................... |
19 |
3 |
13 |
0.62 |
8 |
7 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................GGAGGTGGCGAATCTCAA....................................................................... |
18 |
3 |
20 |
0.60 |
12 |
12 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCCAGGTGGCGAATCTCAA....................................................................... |
19 |
3 |
13 |
0.54 |
7 |
7 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................TCGAGGTGGCGAATCTCAAA...................................................................... |
20 |
3 |
4 |
0.50 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................................TGGCGAGGTGGCGAATCTCAA....................................................................... |
21 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................GGGGTGGCGAATCTCAAA...................................................................... |
18 |
3 |
20 |
0.50 |
10 |
9 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................................................................................................CTAGGTGGCGAATATCAAA...................................................................... |
19 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .............................................................................................................................................................TTTGCTAGGTGGCGAATCTCAA....................................................................... |
22 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TTCGAGGTGGCGAATCTCAA....................................................................... |
20 |
3 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................TAGGTGGCGAATATCAA....................................................................... |
17 |
2 |
10 |
0.40 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................CCGAGGTGGCGAATCTCAAA...................................................................... |
20 |
3 |
5 |
0.40 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCTAGGTGGCGAATCTCAAA...................................................................... |
20 |
3 |
8 |
0.38 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................CTGCTTCCCCTAGCTTG..................................................................................................................................................................................................................... |
17 |
2 |
8 |
0.38 |
3 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
1 |
0 |
0 |
| ....................................................................................................................................................GCTTTCAATTTAGCGAGG.................................................................................... |
18 |
2 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CGAGGTGGCGAATCTCAAAA..................................................................... |
20 |
3 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TGAGAGGTCACGAATATCAA....................................................................... |
20 |
2 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ............................................................................................................................................................CTTTGGGAGGTTGCGAAT............................................................................ |
18 |
2 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCGGGGTGGCGAATCTCAA....................................................................... |
19 |
3 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...............................................................................................................................................................TGCGTGGTGGCGAATCTCAA....................................................................... |
20 |
3 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................GGTGGCGAATATCAAA...................................................................... |
16 |
1 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CCAGGTGGCGAATCTCAAA...................................................................... |
19 |
3 |
16 |
0.25 |
4 |
3 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ..................................................................................................................................................................TAGGTGGCGAATATCAAA...................................................................... |
18 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCCAGGTGGCGAATCTCAAA...................................................................... |
20 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................GGTGGCGAATCTCAAAGAAG.................................................................. |
20 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................GGTCGCGAATCTCAAA...................................................................... |
16 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................ACGAGGTGGCGAATCTCAAA...................................................................... |
20 |
3 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................CCTGCTGCCCCGAGCTTG..................................................................................................................................................................................................................... |
18 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................CCTGCTTCCCCTAGCTTG..................................................................................................................................................................................................................... |
18 |
2 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GGGAGGTGGCGAATCTCAA....................................................................... |
19 |
3 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCGAGGGTGCGAATCTCAA....................................................................... |
19 |
3 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................TGGGTGAATCACGAGAA..................................................................................................................................................................... |
17 |
2 |
16 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................GAGGTGGCGAATCTCAAAA..................................................................... |
19 |
3 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................CAGGTGGCGAATCTCAAA...................................................................... |
18 |
3 |
20 |
0.10 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
| ...................................................................GCGGGTGAATCACGA........................................................................................................................................................................ |
15 |
1 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................TGGCGAATCTCAAAGAGTA................................................................. |
19 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................CGAATCTCAAAGGATAC................................................................ |
17 |
2 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................GCGAGTTGGCGAATCTCAA....................................................................... |
19 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................CTTGCTGCCCCTAGCTTGG.................................................................................................................................................................................................................... |
19 |
3 |
13 |
0.08 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................TGAGGTGGCGAATCTCAAA...................................................................... |
19 |
3 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................GGAGGTGGCGAATCTCAAA...................................................................... |
19 |
3 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................TGGCGAATCTCAAAGGATA................................................................. |
19 |
3 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................GGTGGGTGAATCACGAGAAA.................................................................................................................................................................... |
20 |
3 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CGATGTGGCGAATCTCAAA...................................................................... |
19 |
3 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................CTAGGTGGCGAATTTCAAA...................................................................... |
19 |
3 |
16 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................ACTAGAGCCAAGTTCAT.................... |
17 |
2 |
18 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| .....................................................................GGGTGAATCACGAGAGATA.................................................................................................................................................................. |
19 |
3 |
18 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................TGCTTTCAATTTAGCGA...................................................................................... |
17 |
2 |
19 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................TTAAGAGTAGGCTTGTGG................................................................................................................... |
18 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................TGGCGAATCTCAAAGA.................................................................... |
16 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...........................................................................AACAAGAGAGGAATAAAAACAA......................................................................................................................................................... |
22 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
| .....................................................................CGGTGAATCTAGAGTAAAA.................................................................................................................................................................. |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| GTGTTCAAGTTGAGTCT......................................................................................................................................................................................................................................... |
17 |
2 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ....................................................................................................................................................................GGGCGCGAATCTCAAA...................................................................... |
16 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |