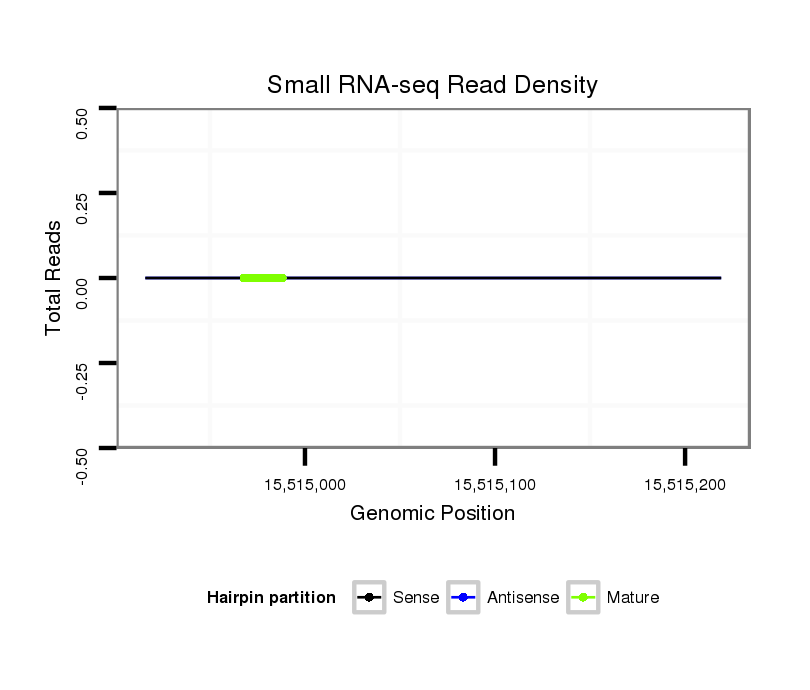
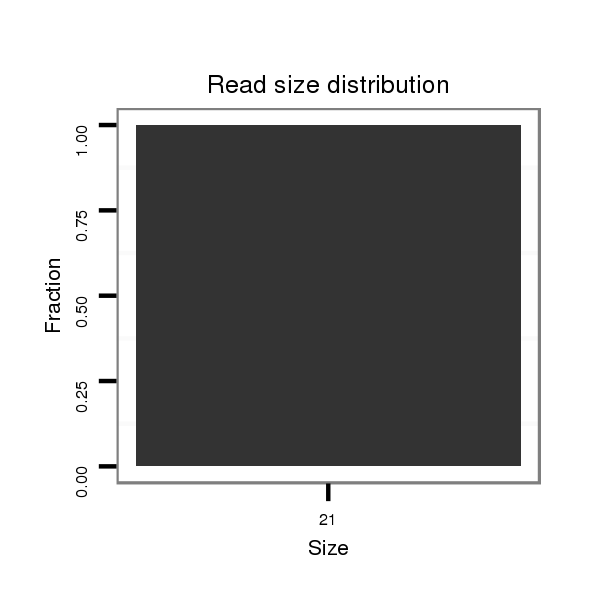
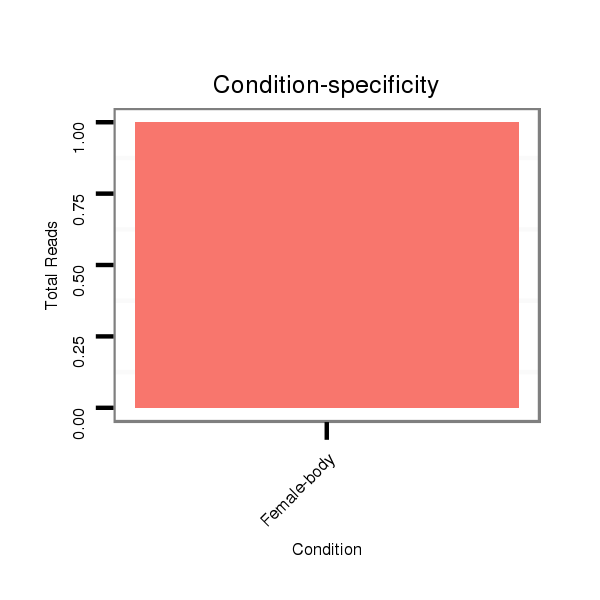
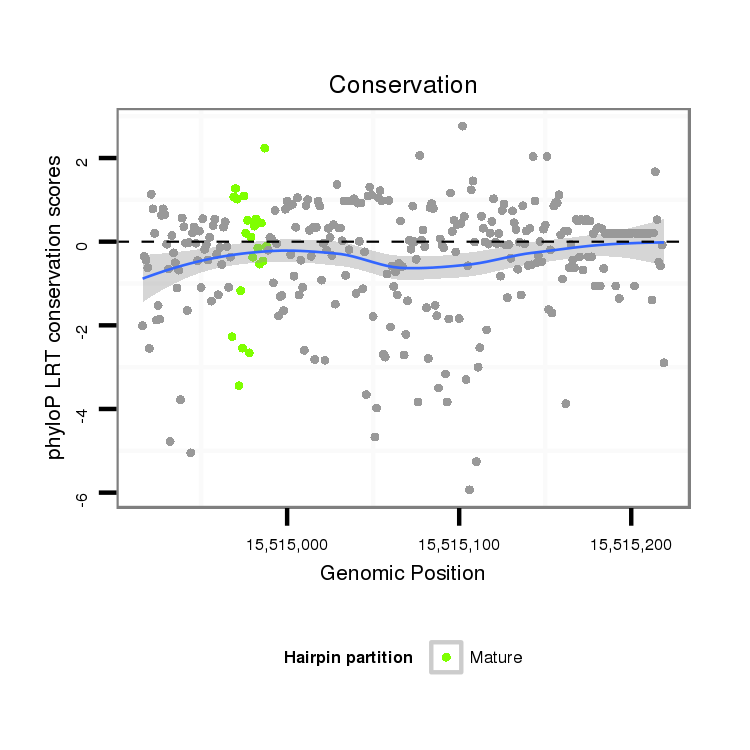
| droSim2 |
2r:15514916-15515219 + |
dsi_24680 |
AG----GTCCAGG-TCC------AGGCCCAGGCTCAGGCTCAGGCCCAGGCCCAGGCCCAGGTTCAGGCTCAGGCTCAGGCTCAGGCTCAGGTCCAGGCTCAGGTCC------------------AGGCTCAGGTC------------------------------------C------------AGGCTCAGGCCCAGGT---------------------------------------------------------------TCAG----------------------------------------------------------------------------------GTTCAGGATCAGGCCCAAGTTCAGGATCAGGCCCAG-------------------------------------------------------------------------------------------------------------------------------------------------GATCAGCTCCAGGGTC------AGGCCCAGGC-CCAGGCCCCGGCTCAGGCCCAGGCCCAGGTCCCGGCTCAGGCCCAG---------------GCCCAGGT---------------------------------CCCG----GC-----------------------TCAGGTCCAGCAGTCATCAGCTCAGTCTGCCCAGCTACTCCTCCCTGA----------------------------------------------------------------------------------------------------------------------------------------GCTA----AC |
| droSec2 |
scaffold_1:12372228-12372532 + |
|
AG----GCCCAGGCCCC------AGGCCCAGGCCCAGGCCCAGGCCCAGGCCCAGACCCAGACCCAGACCCAGACCCAGACCCATACCCAGACCCAGGCCCAGGCCC------------------AGGCCCAGGCC------------------------------------C------------AGGTCCAGGCCCAGGT---------------------------------------------------------------CCAG----------------------------------------------------------------------------------GCCCAGGTTCAGGTCCAGGCTCAGGACCAGGTTCAG-------------------------------------------------------------------------------------------------------------------------------------------------GATCAGCTCCAGGGTC------AGGCCCAGGC-CCAGGTCCCGGCTCAGGCCCAGGCCCAGGTCCCGGCTCAGGCCCAG---------------GCCCAGGT---------------------------------CCCG----GC-----------------------TCAGGTCCAGCAGTCATCAGCTCAGTCCGCCCAGCTACTCCTCCCTGA----------------------------------------------------------------------------------------------------------------------------------------GCTA----AC |
| dm3 |
chr2R:14866123-14866389 + |
|
AG----GCTCAGG-TCC------AGGCACAGGCTCAGGCCCAGGCCCAGACTCAGGCTCAGACTCAGGCCCAGGCTCTGGCTCAGGCCCAGGCTCAGGCCCAGGTCC------------------AGGCACAGCCACAGGCTC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGGTCCAGCAGGCAGCAGCTCAGCCTGCCCAGCTGCTCCTCCCTCAGCTAACGCAGGCGGTGATCCAGACTCAGCAACGAGCGGTTCACCAACCCCAGCAGTCGGGACAGCAATCAGGACCTCAGCATGCCTCATCGAC---CGCCGCC-----------------------------------TT-GGCTA |
| droEre2 |
scaffold_4845:19817866-19818138 + |
|
GG-----CACAGG-CGC------AGGCACACGCACACGCACATGCT---------------------------------------GCCCAGGTACAGGCCCAAGCTC------------------AAGCCCAGGCC------------------------------------G------------TGGTCCAAGCTCAGGTGCAAGCCGCCCAAGTGGCACACGCTGTGGCAGCACAGCAGCAGGCGGTA------------CA-------------------------------------------------------GCAGCAACAGGCGGCACAGCAGCA------------------------GCAGCAAGCCGCGGTGCAA----------------------------------------------------------------------------------------------------------------C---------------------------------AGCAGGCAGCGGTGC------------AGCA-GCAGCAACAG---------------------------------------------------CAACAGGCGGCGGT---------GCAGCAACAGCAG---------------------------------------------------GCAGCG------------------------------------CAGCAGCAGGTCA------AGCAACTGGAGGAACT------------------------------------------------G-----------------------------------------------------AA |
| droYak3 |
X:19876624-19876908 + |
|
AG----CCACAGC-TCC------AGCCTCAGTCACAGCTTCAGCCTCAGTCACAGCCACAGCTTCAGCCTCAGCCACAGTCTCCGGCTTTTCCTCAGTCTTGTCCAC------------------AGCCATAGCCACAGTCTCGACCTC------------AGCCACAG----------------------------------------------------------------------------------------------------------------------------------------------------------------TAGCCACAGCCACAGTCTCGACCTCAGCCGCAGTAGCCGCTGTAGCCTCAGTCACAG----------------------------------------------------------------------------------------------------------------C---------------------------------CTCAGCCACAGTCGC------AGCCTCAGCC-ACAGTCACAGCCTCAGCCACAGTCACAGCCTCAGCCACAGTCACAGCCTCAGCCACAGTCA------------------------------------------CAG----CC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAGCCAC |
| droEug1 |
scf7180000409759:228612-228893 + |
|
AG----TCTCAGT-CTC------AGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCA-----AGTCTCAGTCTCAGTCTC------------AGTCTCAGTCTCAGTCTCAGTCTC------------AGTCTCAGTCTCAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAG----------------------------------------------------------------------------------------------------------------T---------------------------------CTCAGTCTCAGTCTC------AGTCTCAGTC-TCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTCTCAGTTT------------------------------------------CAG----TC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAGTC-T |
| droBia1 |
scf7180000301468:913003-913259 + |
|
TG----GTCCAGG-CCC------AGGTGCAGGCCGTCCAAGTGGCCCACGCTGTGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCCCAGCAGCAGGCGGTG------------CA-------------------------------------------------------GCAGCAGCA------ACAGCAGCA---------GCAGC------------------A-------------------------------------------------------------------------------------------------------------------------------------------------AGCGGCGCAGCAGCAGCAGCAGACGG------TGGCGCAGCA-GCAGCAACAGCAACA------------------------------------------------ACAGGCCGTGGC---------GCAGCAACAGCATG-----------C--------GGCT-----------------------------------------------------------------CAGCAGCAGCAGGTCA------AGCAACTGGAGGAACT------------------------------------------------GAATCGGGCTGTGATGCTCCAGCGCTTTCAGGCTGCCCGGCAGCCGGCAA----AC |
| droTak1 |
scf7180000415191:84658-84961 - |
|
AG----GCCCAGC-AGC------AGCAGAGGGAGCAGCAACAGCAGGTCGCCCAGGCTCAGGCGCAGCAGCAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGGCACTCGCCAACGCCACTCA----GCAGATCCTCCAGGTGGCGCCCAACCAGTTCATCACCTCCCACCAGCAGCAG------------------------------CAGCAGCAGCAGC---TCCACAACCA-------------------------------------------------------------------------------------------------------------------------------------------------ACTGATCCAGCAGCAGCTGCAGCAAC------AGGCGCAGGC-GCAGGCACAGGTTCAAGCCCAGGTGCA---------------------------------------GGCCCAGGC---------GCAGCAGCAACAACAGCAGCGG----GAGCAGCAGCAG-----------------------------------------------------------------CAGCAGCAGCAGAACATCATCCAGCAGATTGTGGTGC--------------------------------------------------------------------------------------------------------A |
| droEle1 |
scf7180000491011:652921-653226 + |
|
GG-----CCCAGT-ACC------AGGCGCAGCAGCAGCAGCAGGCTCAGGCCCAGGCCCAAGCTCAGGCGCAGGCCCAGGCGCAGGCA------CAGGCGCAGGCAC------------------AGGCCCAGGCC------------------------------------C------------A----------------------------------------------------------------------------------GCAGCTAGCC------------------------------------------CAGCAGCAGCTGG------------------CCCAGCACCAGCAGCTGGC------CGC-------------------------------------------------------------------------------------------------------CGCCGTCCAAGTGCATC---------------------------------ACC------------------AGCAGCAGCA-ACAGCAGCAGCA---------------------------------------------------ACAGGCGGTCGTGGCCGTTCAGCAGCAACAGCAGG-----CCGCAGC----AGCGGCGGCTGCGGCGGCGGTCAAGATGCAACT------------------------------------------------------------------------------------------------------------------------------GAC---CGCCGCCACGCC-C---ACCTTCAC---GTTCAGCGCCCTGCCA----AC |
| droRho1 |
scf7180000777112:15271-15523 - |
|
AG----GCTCGGG-ATT------GAGCTCTGGTTCAGGCACTGGTTCGTGTTCCGGTTCGTGTTCCGGATCGGTTTCAGGCTCGGGTTCAGGTTCAGGTTCAGGTTC------------------AGGTTCAGGCT------------------------------------C------------TGGTTCAGGCTCTGGT---------------------------------------------------------------TCAG----------------------------------------------------------------------------------GCTCGGGCTCAGGCTCAGGCTCTGGTTCAGGCTCTG-------------------------------------------------------------------------------------------------------------------------------------------------GTTCAGGCTCTGGTTC------ATGACAAGGCGTCTGGTTCAGGCTCAGGCTCTGGTTCAGGTTCCGGCTCAGGATCTG---------------GATCAGGC---------------------------------TCT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCTC |
| droFic1 |
scf7180000454078:671952-672211 + |
|
AA----GCCCAGG-TGC------AGGCGGCCCAGGTGGCCCACGCTGTGGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGCCCAGCAGCAGGCAGTGGCG---------CA-------------------------------------------------------GCAGCAACAGGCGGCAGTGCAGCA---------GCAGCAACAGCAACAGGC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCAGTGC------------AGCA-GCAGCAACAG---------------------------------------------------CAGCAGGCCGTGGT---------TCAGCAACAACAACAACAGGCG----GCGGTGGCTCAG-----------------------------------------------------------------CAGCAGCAGCAGGTCA------AGCAACTTGAGGAACT------------------------------------------------GAATCGGGCCGTTATGATCCAGCGCTTACAGGCTGCCCGGCAGCCGGCGA----AT |
| droKik1 |
scf7180000302476:2954895-2955252 - |
|
CC--AAAACCAGG-CCCAGATTCAGGC------TCAGGCGCAGGCCCAGGCTCAGGTTCAAGCTCAGGCTCAGGCACAAGCTCAGGCT------CAGGCTCAAGCAC------------------AGGCTCAGGCC------------------------------------A------------ACATGCAG-----------------------------------------------------------------------------------CG------------------------------------------------------------------------------------------------------------------TATCAACTACGCTTATAGCTATGGCAACTTGTATAACCCTTATGGCGGGGTCACCTCCTCCATGGTAAGCCTCACCAGCTCCAGTCCCTCCTACGCCCAGGCACAGATGCAACAGCAGCAGCAGCAGCAA---GCGCAGCAGCAGCAACAAGTGC------AGGCTCAAGT-CCAGGCCCAGGCTCAGGCC------------------------------------------------GCCTATGC---------CCAACAGGCCTAT-----------GC----AGCCTATG-----------------------------------------------------------------CTGCGGCTGCGGGACT------AGCTCCCCAAGCCAC--------------------------------------------------------------------------------------------------------T |
| droAna3 |
scaffold_12916:14486840-14487150 + |
|
AA----GCCCAGG-CTC------AAGCTCAAGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAAGCTC------------------AGGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCAAGCTCAGGCTCAAGCTCAGGCTCAAGCTCAG----------------------------------------------------------------------------------------------------------------G---------------------------------CTCAAGCTCAAGCTC------AGGCTCAAGC-TCAGGCTCAAGCTCAAGCTCAAGCTCAAGCTCAGGCTCAAGCTCAGGCT--------------------------------------------------------TCAGC----TTCGGCTTCAGCT---------------------------------------------------------------CAA-----------------------------------------------------------------------------------------------GCT-C---AGGCCCAA---GCTCAGGCTCCAGCTCAAGC-T |
| droBip1 |
scf7180000396572:2493016-2493319 - |
|
AA----GCACAGG-CTC------AAGCTCAAGCACAGGCTCAGGCACAGGCTCAAGCTCAAGCACAGGCTCAAGCTCAAGCACAGGCTCAAACACAGGCTCAGGCACAGACTCAAGCACAGGGTCAGGCTCAGGCG------------CAGGCTCAAGCACAGGCTCAAGCTCAGGCTCAAGCTCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTCAAGCACAGGCTCAAGCTCAGGCTCAGGCGCAG----------------------------------------------------------------------------------------------------------------G---------------------------------C------------TC------AAGCTCAGAC-ACAGGCTCAAGCTCAGGCTCAAGCTCAGGCACAGGCTCAAGCTCAGGCACAGGCTCAAGCT------------------------------------------CAG----GC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCAG---GCTCAAGCTCAGGCTCAGG--C |
| dp5 |
XL_group3a:1834191-1834499 - |
|
GG-----CACAGG-CTC------AGGCCCAAGCTCAGGCCCAAGCTCAGGTTCAGGCTCAGCAACAGCAGCAG------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCAGCAGGCATTGGCG---------CA----GCAGATACTGCAGGTGGCGCCAAACACCTTCATCACCTCCCACCA---------------------ACAGCAGCA------GCAGCAGCAGCAGC---TCCACAACCA-------------------------------------------------------------------------------------------------------------------------------------------------ACTGCTGCAGCAGCAGCTCCAGCAGC------AGGCGCAGGC-CCAAGTGCAAGCTCAGGTTCAGGCTCAGGCACAGGCACAGGCCCAG------------------------------------------CAGCAACAGCAACAGCAACAGC----AGCGGGAG-----------------------------------------------------------------CAGCAGCAGCAGAACATAATCCAACAAATTGTGGTAC--------------------------------------------------------------------------------------------------------A |
| droPer2 |
scaffold_4:5078269-5078555 - |
|
TG----GCTCACT-CTT------AGGTTCTGGCTCACTCTTCGGCTCTGGCTCACTCTTCGGCTCAGGTTCGCTCTGCGGCTCAGGGACGCTCTTCGGCTCAGGGAT------------------GCTCTTCGGCTCAGGTTCGCTCTT------------AGGTTCACTCTTAGGTTCTGGCTCAC-------------------------------------------------------------------------------------------------------------------------------------------------------------------CCTTAGGTTCAGGTTCGCTTTTAGGTTCTGGTTCGC----------------------------------------------------------------------------------------------------------------T---------------------------------CTTCGGCTCGGGCTCACTCTTCGGCTCAGGC-TCACTCTTCGGCTCAGGTTCACTCTTCGGCTCAGGCTCGCTCTTCGGTTCTGGTTCGCTCT------------------------------------------TCG----GC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAGGC-T |
| droWil2 |
scf2_1100000004943:10293260-10293473 + |
|
GGCT--CGGGTTC-TTC------TGGAGCGGGTTCTGGCTCAGGTTCTGG------------CACAGGTTCGGCCTCAGGTTCTGGCT------------------------------------------------------------------------------------C------------AGCTGCGGGCACAGGT---------------------------------------------------------------TCTG----------------------------------------------------------------------------------GCTCAGGTACTGCCTCAGGCTCTGGCTTAGGTTCTGGAACTATTTCCACTA-------------------------------------------------------------------------------------------------------------------------------------------CAGCTTC------TGGTTGAGGT-TCAATTTCTAACTT------------TGTCTCCTGTTCAGGCTCAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTCAG---GCTCTGCTTCCGGTTC----AG |
| droVir3 |
scaffold_13042:764977-765256 + |
|
GG----ATCCAAT-TCC------AGCTCCAGTCCCAGTCTTAGTCCCAGTCCTAGTCCTAGTCAAAGCTTTAGTCTCAGTCCCAATCCCAGCTCCAGTCCCAGCTCC------------------AGTCCCAGCTCCAGTCCCAGCTTC------------AGTCCCAGTCCTAGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------CCCAGCCCCAGCTCCAGTTTTAGCTCCTGTCCCAACCCCAGTTGCAG----------------------------------------------------------------------------------------------------------------C---------------------------------CCCAGCCACAGTCCC------AGCTCCAGTC-TTAGCTCCAGTCTTAGCTCTAGCCCCAATCCCAGTCCCAGTCCTAGTCCCAGTCCTTGTCC------------------------------------------CTG----TT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAGT--C |
| droMoj3 |
scaffold_6496:14702555-14702858 + |
|
AT----GCTCAGG-CCC------AGGAACAGGCTCAGGCTCAAGCTCATGCGCAACTTCAGGCCCACGCCCAGCTCCAAGCTCACGCCCACCTCCAGGCCCAAGCTCAAGCTC------------AGGCTCAGGCTCAGGCCCAGGCCCAAGCCCAGCTCCAGGCACAGGTTCAGGCCCATGCCCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------------CTCCAGGCACACGCCCATGCACAGGTTCAGGCCCAG-------------------------------------------------------------------------------------------------------------------------------------------------------------GCCC------AGCTCCAGGC-GCAAGTCCAGGCCCATGCTCATGCCCAGCTCCAGGCGCATGTCCAGGCCCAGGCACACGCA------------------------------------------CAG----GC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAG---GCACATGCTCATGCACAGG--C |
| droGri2 |
scaffold_15074:3889875-3890160 - |
|
CA---GGTCCAGT-CTC------AGGCTTACCTCCAGATACAGCCCCAGCTCCACCTCCAGGTCCAGCATCAGTTTTAATTCCCGGTCCGACGTCAGTCTTATCTCC------------------AGGTCCAGCACCGGGCTTAACTGC------------GGGTCCAGCACC------------AGGTGCAGC-------------------------------------------------------------------------------------------------------------------------------------------------AACAGGTTTAGCTGCAGGATCAGCACTTGGCTTAACAGCGGGTCCAG-------------------------------------------------------------------------------------------------------------------------------------------------CACCAGCTCTACCACC------AGGTCCAGCC-ACAGCTTTAACTCCCGGTTCAGCACCAGTCTTATCTCCACGTCCAGTACCACGTTTATCTC------------------------------------------CAG----AT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAG---C |