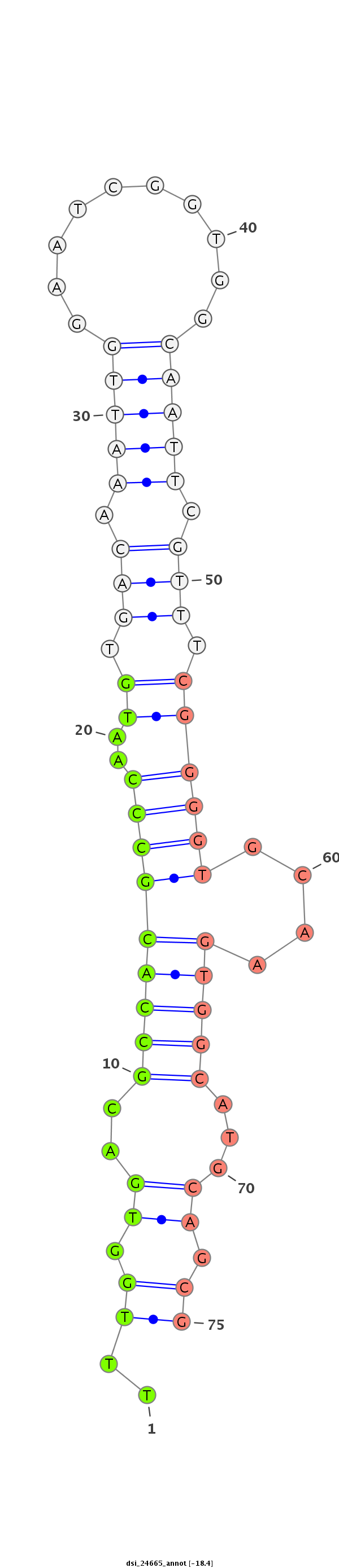
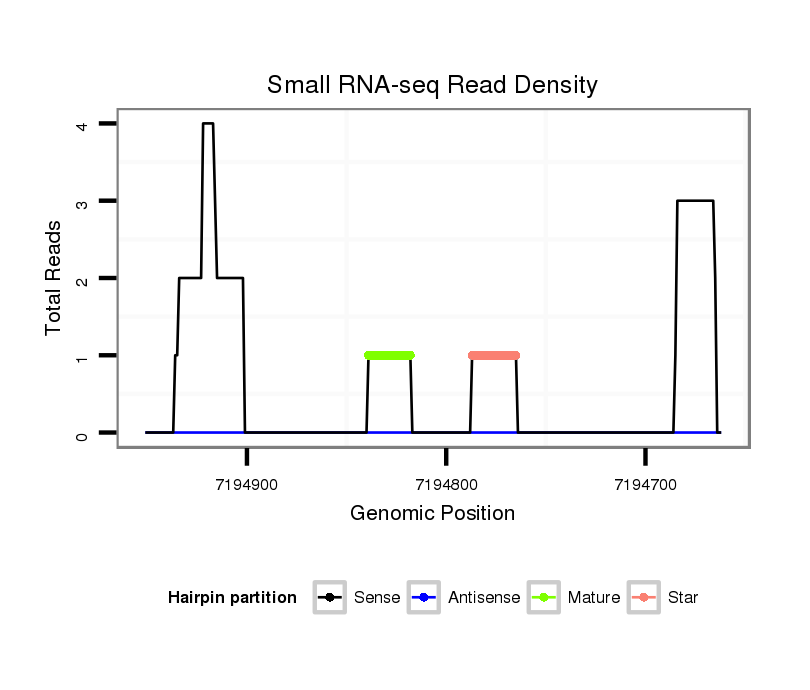
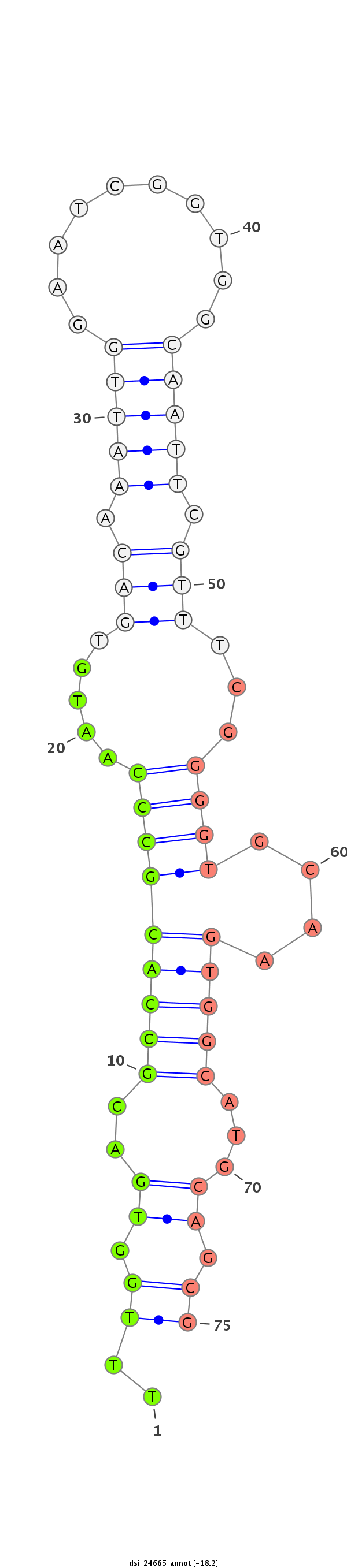
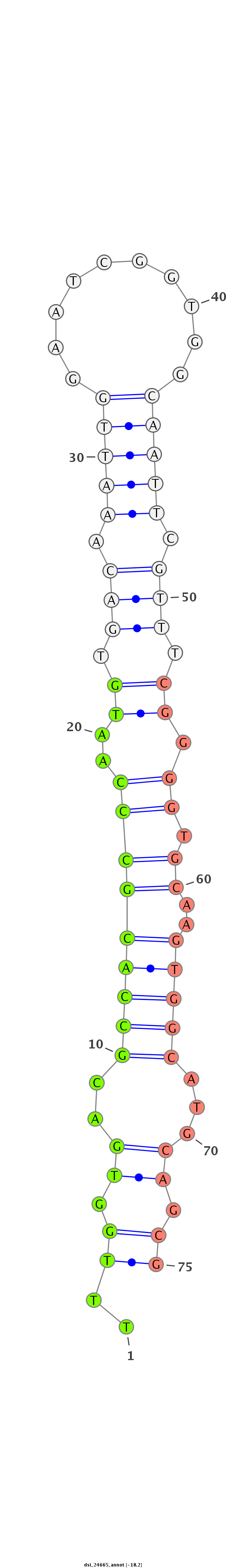
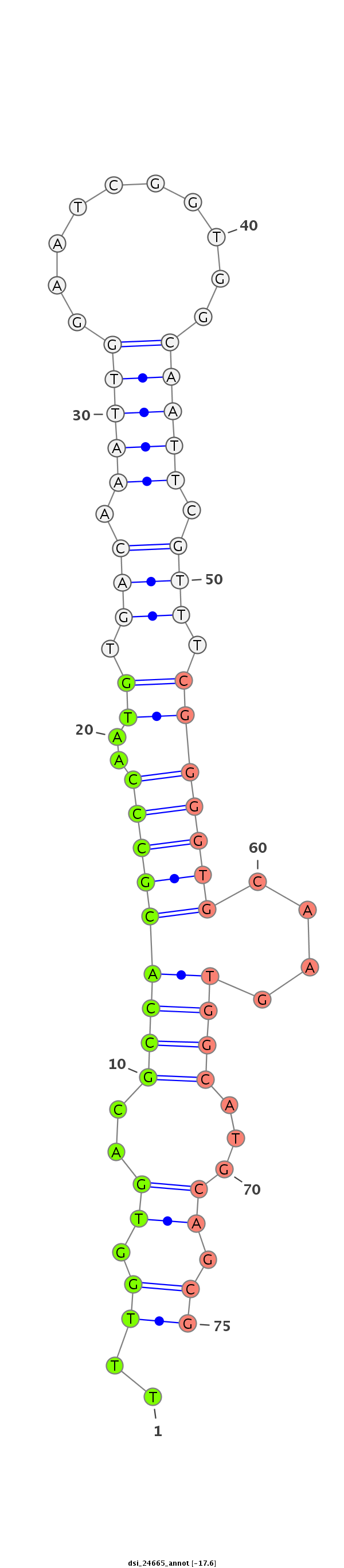
ID:dsi_24665 |
Coordinate:3l:7194712-7194901 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -18.2 | -18.2 | -17.6 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCTGAATGGGACTTGTGCCAATCTCTGTGACCACCAATCGCGCGCAAATGGTAAGTGCAATCCGCATCTAAGCTGCAAAGTGGGGAGCTGCAGGTGCTGCTAGTTAGTTTACTTTGGTGACGCCACGCCCAATGTGACAAATTGGAATCGGTGGCAATTCGTTTCGGGGTGCAAGTGGCATGCAGCGTGGTGGTTGAACCGCTTTCTCATTGCGAACGCATTTCTTCTCCTGCCCTCCAGACCATTGAGCATTGTGCAGCAGGCTATTGAGGCTCTGGCAAGTAACCACA
****************************************************************************************************************..((.((..(((((((((..((.(((.(((((..........))))).))).))))))....)))))...)).))******************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M025 embryo |
M024 male body |
O001 Testis |
O002 Head |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................ACCACCAATCGCGCGCAAATG................................................................................................................................................................................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGAGGCTCTGGCAAGTAAC.... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............TGCCAATCTCTGTGACCACCA.............................................................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TTTGGTGACGCCACGCCCAATG............................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................CGGGGTGCAAGTGGCATGCAGCG....................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................TTGAGGCTCTGGCAAGTAACC... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................CCAATCTCTGTGACCACC............................................................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................TGAGGCTCTGGCAAGTAACC... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................AATCGCGTGCAAATGGT.............................................................................................................................................................................................................................................. | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................ATTCGTTTCGGGATG....................................................................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................GAACAAATTGGAATCGGT.......................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................GCATCTAAGCTGGAA.................................................................................................................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................TCCTGCCGTCCTGACCA.............................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GGACTTACCCTGAACACGGTTAGAGACACTGGTGGTTAGCGCGCGTTTACCATTCACGTTAGGCGTAGATTCGACGTTTCACCCCTCGACGTCCACGACGATCAATCAAATGAAACCACTGCGGTGCGGGTTACACTGTTTAACCTTAGCCACCGTTAAGCAAAGCCCCACGTTCACCGTACGTCGCACCACCAACTTGGCGAAAGAGTAACGCTTGCGTAAAGAAGAGGACGGGAGGTCTGGTAACTCGTAACACGTCGTCCGATAACTCCGAGACCGTTCATTGGTGT
*******************************************************************************************************..((.((..(((((((((..((.(((.(((((..........))))).))).))))))....)))))...)).))**************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................CGCGAGATCTGGTAACTCGG....................................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TTGGCGATAGAGTAA............................................................................... | 15 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................AGGTATGGTAACTCG........................................ | 15 | 1 | 12 | 0.17 | 2 | 0 | 0 | 2 | 0 |
| .....................AGAGACGCTGGTGGT.............................................................................................................................................................................................................................................................. | 15 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .......................................................ATGTTGGGAGTAGATTCGA........................................................................................................................................................................................................................ | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AGAGTAACGCTTGCT....................................................................... | 15 | 1 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................................................................................................................TCCGAGGCAGTTCATAGGT.. | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 |
| .....................AGAGAGACTGGTAGTTA............................................................................................................................................................................................................................................................ | 17 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................AGGTCTCGTGACTCGTA...................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 08:05 PM