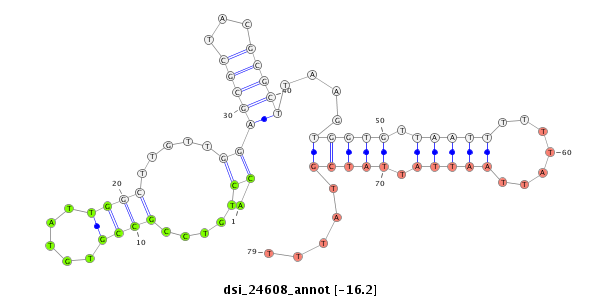
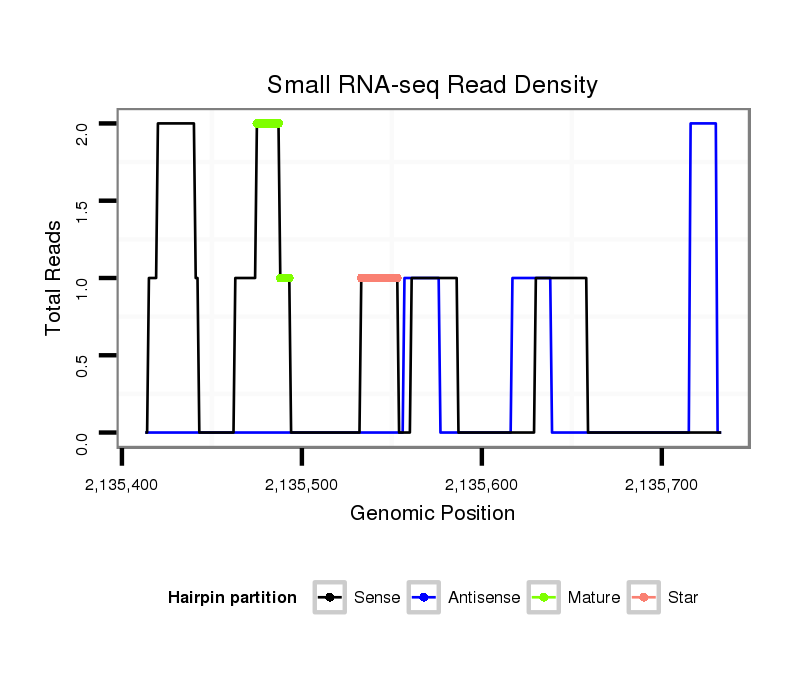
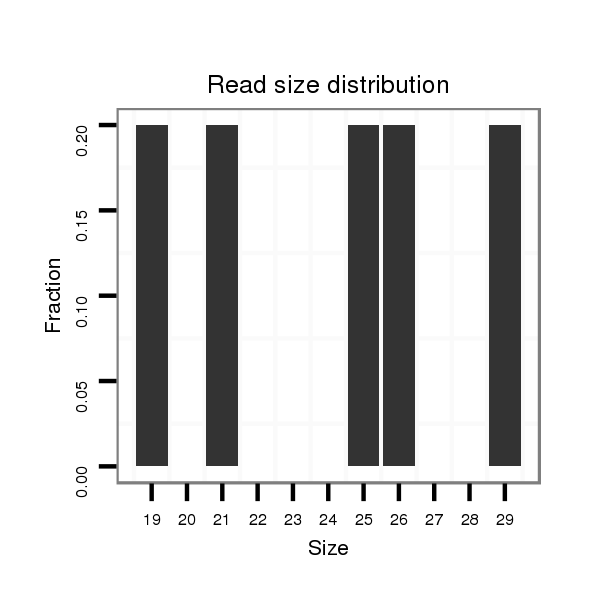
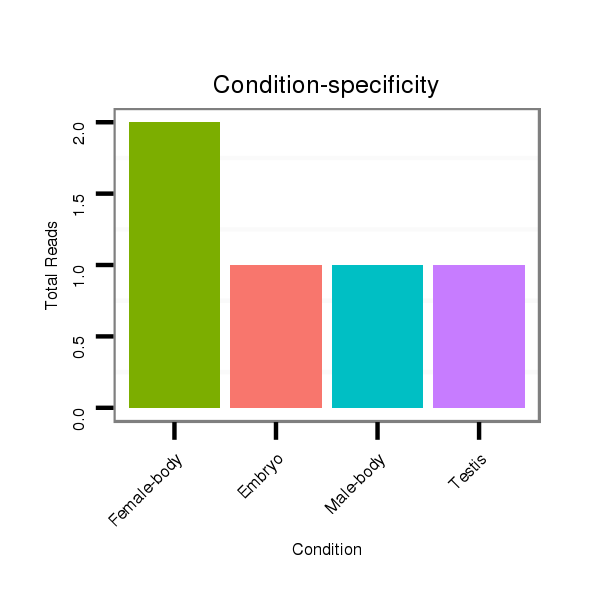
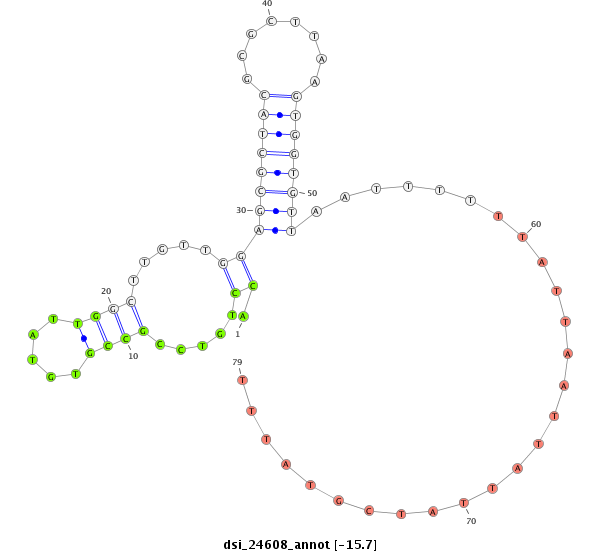
ID:dsi_24608 |
Coordinate:3l:2135463-2135683 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -15.7 | -15.6 | -15.5 |
 |
 |
 |
five_prime_UTR [3l_2135353_2135756_+]; exon [3l_2135353_2135756_+]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ################################################################################################################################################################################################################################################################################################################################# ATTAAGTCGGACAGAATCGAATCGGGGCGAGCGCTTATCGCGGCAAAAGAGTAATTTGTGATACCTGTCCGCCGTGTATTGGCTTGTTGGAGCGCTACGCGCTTAAGTGGTGTTAATTTTTTATTAATTATTATCGTATTTATTACCCAACGAAAGTGAAGTGCGAAATCGGTCGCCTCAAGTGCCCGGCCGTACGTGCAAAGCCCGTAAATAAATTACACAGCCATATAGCCCTGATCCTTACCTCCACACTCGCTCTCGCCCATTGCAGCTGATAATGCAGACGAAGTCCGCCTGATCGATTCCCATTCCCATTTTGAT **************************************************************.((.....((((.....)))).....))(((((...)))))....(((((.(((((.......))))).))))).....************************************************************************************************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
M025 embryo |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..TAAGTCGGACGGGAACGAATC.......................................................................................................................................................................................................................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................ACCTGTCCGCCGTGTATTG................................................................................................................................................................................................................................................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................AGACGAAGTCCGCCTGATCGAC.................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......CGGACAGAATCGAATCGGGGCGA................................................................................................................................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TTATTAATTATTATCGTATTT.................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AACGAAAGTGAAGTGCGAAATCGGTC................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAATTTGTGATACCTGTCCGCCGT...................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .TTAAGTCGGACGGGAACGAATC.......................................................................................................................................................................................................................................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..TAAGTCGGACAGAATCGAATCGGGGC..................................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................ACACAGCCATATAGCCCTGATCCTTACCT........................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................GTACGTGAGAAGACCGTAAA.............................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................GCCGGACGTGCAAGGCTCG.................................................................................................................. | 19 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................CGAAATCGGCCGCTTCAA............................................................................................................................................ | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................CCAAGTCCTCCTGATCGA................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............AATCGACTCGGGGCG.................................................................................................................................................................................................................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................AGTCCTCCTGATCGA................... | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................................................................CGCCGGTTCGATTCCCA............. | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........CAGAATCGGATCGGTGC..................................................................................................................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................TGTTGACGAAGTCCGCC.......................... | 17 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................GCCTGGTAGAGCGCTACG.............................................................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
TAATTCAGCCTGTCTTAGCTTAGCCCCGCTCGCGAATAGCGCCGTTTTCTCATTAAACACTATGGACAGGCGGCACATAACCGAACAACCTCGCGATGCGCGAATTCACCACAATTAAAAAATAATTAATAATAGCATAAATAATGGGTTGCTTTCACTTCACGCTTTAGCCAGCGGAGTTCACGGGCCGGCATGCACGTTTCGGGCATTTATTTAATGTGTCGGTATATCGGGACTAGGAATGGAGGTGTGAGCGAGAGCGGGTAACGTCGACTATTACGTCTGCTTCAGGCGGACTAGCTAAGGGTAAGGGTAAAACTA
************************************************************************************************************************************************************************************.((.....((((.....)))).....))(((((...)))))....(((((.(((((.......))))).))))).....************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................................................................................AGGGTAAGGGTAAAA... | 15 | 0 | 10 | 2.10 | 21 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..ATTCAGCCTGCCCTTGCTTAG.......................................................................................................................................................................................................................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................TTCAGGCGGACTACGTCAGGGT............. | 22 | 3 | 3 | 1.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TGGGTTGCTTTCACTTCACG............................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGCATTTATTTAATGTGTCGGT............................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................AAGGGTAAGGGTAAAA... | 16 | 0 | 9 | 0.78 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................GGTGCACGGGCCGGCATG.............................................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GGTGCCGGTATGTCGGGACT.................................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................AGTGCACAGGCCGGCATG.............................................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................AGCCGGCGGAGTTCACG........................................................................................................................................ | 17 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........CCTGTCTTAGCTTTGC......................................................................................................................................................................................................................................................................................................... | 16 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AGCCGGCAGAGTTCACGGCCC.................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...TTCAGCCTGCCCTTGCTTAG.......................................................................................................................................................................................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................TATTAGGTCTGCTTCAAG............................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................CAGGCTTTGGCCAGC.................................................................................................................................................. | 15 | 2 | 20 | 0.20 | 4 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 |
| ...........................................................................................................................................................................................................................................CCAGGAATGGTGGTTTGAGC.................................................................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................TTTAAATACTATGGACAG............................................................................................................................................................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................AGGGTAAGGGTAAAAC.. | 16 | 0 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................TGCAAGCTTTAGTCAGCG................................................................................................................................................. | 18 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| .................................................................................................................................................................ACGGTTTGGCCAGCGGA............................................................................................................................................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....TCAGCCTGCCCTTGCTTAG.......................................................................................................................................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AGCCGGCGGAGTTCA.......................................................................................................................................... | 15 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................ATGCGCGAATACATCA.................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTCACTTGAGGCTTTAG....................................................................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........GTCTTAGCTTGGCCCA...................................................................................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:2135413-2135733 + | dsi_24608 | ATTAAGTC-----G------GA--CAG------------------AATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACC---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GGCCGTA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGCC-CTGATCCTTACC-------------------------------------------------------------------TCCACA-CT-C--GCT-----------------------------------------------CTCGC-CCATT---GCAG---CTGATAATGCAGA----CGAAGTCCGCCTGATCGATTCCCATTCCCAT---------------------------TT-TGAT |
| droSec2 | scaffold_2:2263416-2263736 + | ATTAAGTC-----G------GA--CAG------------------AATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCA---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACC---CCA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GACCGTA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGTA-CTGATCCTTACC-------------------------------------------------------------------TCCACA-CT-C--GCT-----------------------------------------------CTCGC-CCACT---GCAG---CTGATAATGCAGA----CGAAGTCCGCCTGATCGATTCCCATTCCCAT---------------------------TT-TGAT | |
| dm3 | chr3L:2237670-2237990 + | ATTAAGTC-----G------GA--CGG------------------AATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--AACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACC---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GGCCGTA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGCC-CTGATCCTTACC-------------------------------------------------------------------TCTACA-CT-C--GCT-----------------------------------------------CTCGC-CCACC---GCAG---CTGATAATGCAGA----CGAATTCCGCCTGATCGATTCCCATTCCCAT---------------------------TT-CGAT | |
| droEre2 | scaffold_4784:2234354-2234668 + | ATTAAGTC-----G------CA--CGG------------------AATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACT---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GTCCGTA-----------CGTGC-AAAGCCCGCAAATAAA--------TTACACAGCCATAT--AGCC-GTGGTCCTTCCC-------------------------------------------------------------------TTTACA-CT-C--G------------------------------------------------------CCCACT---GCAG---CTGATAATGCAGA----CGAAGTCCGCCTGATCGATCCCCATTCCGAT---------------------------TC-CGAT | |
| droYak3 | 3L:2192195-2192533 + | ATTAAGTC-----G------CA--CGG------------------AATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACT---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GTTCCTA-----------CGTGC-AAAGCCCGCAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCCC-------------------------------------------------------------------TCTATA-CT-C--GCT-----------------------------------------------CTCGT-CCACT---GCAG---CTGATAATGCAGA----CGAAGTTCGCCTGATCGATTCCCATTCCCATTCCTATTCCG---ATTCCGA---T---TC-GGTT | |
| droEug1 | scf7180000409091:26556-26873 - | GTTAAGTC-----G------GA--CGA-AAC------------GAAATCG----------GAATCGGGG--CGAGCG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCAGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACT---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--G-------GGCCATA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCCC-------------------------------------------------------------------TTTACC-CT-T--A------------------------------------------------------CCCACCGCCGCAG---CTGATAATGCAAA----CGGAGTCCGCCTGATCGATTT------CGTT---------------------------TT-GGAG | |
| droBia1 | scf7180000302428:8971643-8971971 + | ATTAAGTC-----G------CC--TGA------------------AATCGAAAT----------CGGGG--CGAGCGCCTTATCG----CGGGTGA---AA-----AAAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTTCGC----CG-AGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACT---CAA--CGAAAGTGAAGTGCC-A---AAT-------------------------------CGGCCG--CCTCAAGTGCC--C-------GGCCATA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACCCAGCCATAT--AGCC-CTGATCCTTGCC-------------------------------------------------------------------TC----TTT----------------TCCGCCCT-----TCCATT-----------CCGGCACT------CTACT---GCAG---CTGATAATGCAAA----CGGACTTCGTCTGATTA--------------------------TAT------------TT-CGAT | |
| droTak1 | scf7180000414400:203078-203423 + | GTTAACTC-----G------CT--GGG-----------------AAATCGAAAT----------CGGGAACGGAACGCCTTATCG----CGGCAGA----------GAAAAGAGTAATTTGTGATACCTGTCCGCCGTGTTGTATTGGCTTGTT-GA----AG-CGCTCTACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACC---CAACGTGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--A-TTCGTTGGCCATA-----------CGTGC-AAAGCCCATAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCC--------------------------------------------------------------------TC----TTTTT---------------C-----CCCACTTGTATTCCA-----TTCG-------------AAACT---GCAG---CTGATAATGCAAA----CGGACTCCGTCTGATCGATTTA---------------------TAT------------AT-TGGC | |
| droEle1 | scf7180000491249:3230475-3230772 - | CCTCACTC-----G------AA--CGG-ACG------------GGAATAC---TTATGAGGAATCGGGG--CGAGTG-CTTATCG----CGGAAGAAG-AA-----AAAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GA----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT--TTATTAATTATTATCGTATTTATT-ACC---CAACGTGAAAGTGAAGTGCA-A---AAT-------------------------------CGCACGGTGCTCAAGTG----------------------------------C-AAAGCCCGTAAATAAA--------TTACACTGCCATATATAGCC-GTGATCCTTTTC---------------------------------------------------------------------------------------------------------------------------CGTT--------CAAATT---GCAA---CTGATAATGCAAA----CGGACTCCGTCTGA--------------------------------------------------- | |
| droRho1 | scf7180000779532:40807-41113 - | GCTCACTC-----G------CA--CGG------------------AATCGA-AT----------CGCGG--CGAACG-CTTATCG----CGGG--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACC---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGCTCGG-CCTCAAGTGCC--C-------GGCCATA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCTT-----------------------------------------------------------------CCCC----TTTTGCAACC-CACTCATC-------------------------------------------------------A---CTGATAATGCAAG----CGGACTCCGCCTGATCGAT---------------------------------------TT-GGAG | |
| droFic1 | scf7180000453052:817980-818302 - | CCTATATCTTCACCCACGG-GG--CGG------------------AATCGA-AT-----CGAATCGGGG--CGAGTG-CTTATCG----CGGC--------------AAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-ACT---CAA--CGAAAGTGAAGTGCG-A---AAT-------------------------------CGGTCG--CCTCAAGTGCC--C-------GGGCATA-----------CGTGC-AAAGCCCGTAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCCCCCATTCC------------------------------------------------------ATT-----G-----------------------------------------CCAATCCGATT-------------CCGACT---GCAG---CTGATAATGCCAACAA-CGGACTCCGCCTGAT-------------------------------------------------- | |
| droKik1 | scf7180000302441:1548915-1549215 - | ATT--GTG-----G------GA--CGG------------------AATCGA-AT-----GGAATCGGGG--CGAGCG-CTTATCG----CGGTGGCAGCAAACAGCAAAAAGAGTAATTTGTGATACCTGTCCGCC---GTGTATTGGCTTGTT-CG----AG-CGC--TACGCGCTTAAGTGGTGTTAATTT-TGTATTAATTATTATCGTATTTATT-GGC---CAGCGTGAAAGTGAAGTGCT-G---AAG-------------------------------CA--AG--CCTCAAGTGCA--G-------CGCCATA------------------GAGCCAATAAATAAA--------TTACACGGCCATATAAAGCCAGTGATCCTTTGC--------------------------------------------------------------------------------------CTGAT--C-----CCCAGTTG----CCA-----TTC-------------CAAACT---GCAG---CTGATAATGCTG---------------------------------------------------------------------- | |
| droAna3 | scaffold_13337:600137-600418 - | GTTACGGG-----G------GA--CGGCGAC------------GAAATCGA-AT----------CGGGG--CGAGCG-CTTATCG----CGCCAGA----------AAAAAGAGTAATTTGTGATACCTGTCC-CC---GAGTATTGGCTTGTT-GG----AG-CGC--TACGAGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATT-GGC---AAC--CGAAAGTCGAGTGCG-A---AAA-------------------------------CGG------CTCAAGTGCTCTC-------GGC---------------CGTGC-TAAGCCAATAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCCC-------------------------------------------------------------------A-------------------------------------------------------------------CCAATT---GCAG---CTGATAATGCACA----CGGACTTT-------------------------------------------------------AT | |
| droBip1 | scf7180000395155:160524-160813 + | GTAGCGGG-----------GTA--CGGCGAC------------GAAATCGA-AT----------CGGGG--CGAGTG-CTTATCG----CGCCAGA---AA-----AAAAAGAGTAATTTGTGATACCTGTCCGTT---GAATATTGGCTTGTT-GG----AG-CGC--TACGCGCTTAAGTGGTATTAATTT-TTTATTAATTATTATCGTATTTATT-GCC---AAC--CGAAAGTGGAGTGCG-A---AAA----------------------------------ACG--ACTCAAGTGCTCTC-------GGCCATA-----------CGTGC-TAAGCCAATAAATAAA--------TTACACAGCCATAT--AGCC-GTGATCCTTCCC-------------------------------------------------------------------A-------------------------------------------------------------------CCAATT---GCAA---CTGATAATGCACA----CGGACTTT-------------------------------------------------------AT | |
| dp5 | XR_group6:12159090-12159439 + | ACTGGC-------------GTAAGCCTCAAC------------GAGCAGCA-AT----------C-------GACCG-CTTATCAGCCCTTGC--------------CAAAGAGTAATTTGTGATACCTGTCCGCCTT-GTGTATTGGCTTGTTTAG----AG-CGC--AACGCGCTTAAGTGGTGTTAATTT-TTCAGTAATTATTATCGTATTTATT-GCC---GAA--CGAAAGTGAAGTGCG-ATCGAACCCTC---------CGCTTCCTCCGCCCCCACCGCTC---CGTCCAGTGCC--G-TGT---GTCCGTC-----------CGTGCGAATGGCAATAAATGAA--------TTACAGAGCCATAA--AGTT-GT------------------------------------------------------------------------------------------------CTGATCAGCGTGGT-----CGGGTT-----------GCGGTCGTCTCGGCCAACT---GCAG---CTGATAATGGACA--TGGGAAGTCTACGTG------------------------------------------TGTG--TGCA | |
| droPer2 | scaffold_33:182229-182587 + | ACTGGC-------------GTAAGCCTCAAC------------GAGCAGCA-AT----------C-------GACCG-CTTATCAGCCCTTGC--------------CAAAGAGTAATTTGTGATACCTGTCCGCCTT-GTGTATTGGCTTGTTTAG----AG-CGC--AACGCGCTTAAGTGGTGTTAATTT-TTCAGTAATTATTATCGTATTTATT-GCC---GAA--CGAAAGTGAAGTGCG-ATCGAACCCTCCGCTGCCTCCGCTTCCTCCGCCCCCACCGCTC---CGTCCAGTGCC--G-TGT---GTCCGTC-----------CGTGCGAAAGGCAATAAATGAA--------TTACAGAGCCATAA--AGTT-GT------------------------------------------------------------------------------------------------CTGATCAGCGTGGT-----CGGGTT-----------GCGGTCGTCTCGGCCAACT---GCAG---CTGATAATGGACA--TGGGAAGTCTACGTG------------------------------------------TGTG--TGCA | |
| droWil2 | scf2_1100000004729:1124403-1124644 - | G-------------------------------------------------------------------------------------------C--------------CAAAGAGTAATTTGTGATACCTGTCCGTTATATTGTATTGACTTGTA-GC----AGTCGC--AACGAGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTATTTATTTAACCA-GAA--CGAAAGTGAAGTGCG-A---ATT--------------------------------G--CA--TTTCAAGTGTT--T-------CGACATTAACCTAACTGGCATG--AAAGGCAATAAACAA---------TTGTG---------------------------------------------------------------------------------------------CATC----GGT-G--G-TC---T-----------------------------------------------GCAATG---GCAG----AGATAAAGTAAT----CAAACACC--------------------CAA---------------------------GC-TGGC | |
| droVir3 | scaffold_13049:11872643-11872979 - | GCTCAGTT-----G------AG--CAGCAATAGCCGTAACCGT-CGCTGG----------CAATCGA----CG-ACG-CTTATCAGC-TTTGC--------------CAAAGAGTAATTTGTGATACCTGTCCGTT---TTGTATTGGTTTGTA-GC----AG-CGC--GACGAGCTTAAGTGGTGTTAATTT--TTATTAATTATTATCGTGTTTATT-GC----CAA--CGAAAGTGAAGTGCGTG---AAT-------------------------------T---TA--TTTCAAGTGGT--TGTG----TGTTATTAAC--------CAGGC-AACCAC----AATAAATAAC----ATATCTATCCA-AA--ATCC-AAGAACAATCA--------------------------------------------------------------ATT-----G---------------------CAACACGGT-----CCAG----------TTTG-------------CAACT---GCAGCCACAGATAGGGCTTT----ACAAA------------------ATTCCTA------A-CAGGGTCT------------ATTGGTG | |
| droMoj3 | scaffold_6680:8956982-8957280 + | ---------------------------------------------------------------------------CG-CTTATCAGC-TTTGC--------------CAAAGAGTAATTTGTGATACCTGTCCGTT---TTGTATTGGGTTGTA-GC----AG-CGC--GACGAGCTTAAGTGGTGTTAATTTTTTTATTAATTATTATCGTATTTATT-GC----CAA--CCAAAGTGAAGTGCG-A---ATT------------------------------------A--TTTCAAGTGTT--T-------TGCCATTAA-------------------CC----AATAAA--------TTACAAT-----A----CGC---GAAC-------------CAAACAAATTGCACATTACAAAATAACAACAACAATAAAGAAGCGAAAGAAGCAATTGCATC----GCT-C--GCTG---TGA---------------CCAG----------TGGG-------------C--------CAG----AGATAATA-----------------C-------GTTTTCGTACAAAT---------------------------TT-T-TT | |
| droGri2 | scaffold_15110:11318791-11319092 - | ---------------------------------------------------------------------------CG-CTTATCAGC-TTTGC--------------CAAAGAGTAATTTGTGATACCTGTCCGTT---TTGTATTGGCTTGTA-GAGCGCAG-CGC--GACGAGCTTAAGTGGTGTTAATTT-TTTATTAATTATTATCGTGTTTATT-GCC-AGCAA--CGAAAGTGAAGTGCG-A---ATT------------------------------------A--CTTCAAGTGCTC-T-------GTCCATTAA-------------C-CATTCCAGTAAC-AA-----TAAATTACAAA---------AGTT-AAGAAC---------------------------------------------------------------AGCAGCTGCCTT----TTT-T--G---------------TTGT-----CCAG----------TTTG-------------CAAAT---GCAACCTCAGATAATGGTTA----AGAAATTT------------TCAATTTCTCT----ATATA---TATATGTATGTGTG----TGTG |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/19/2015 at 01:48 PM