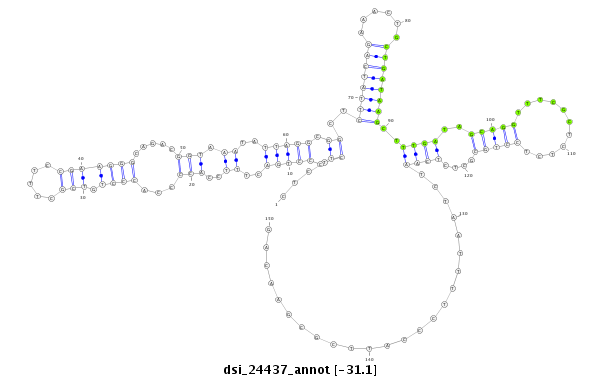
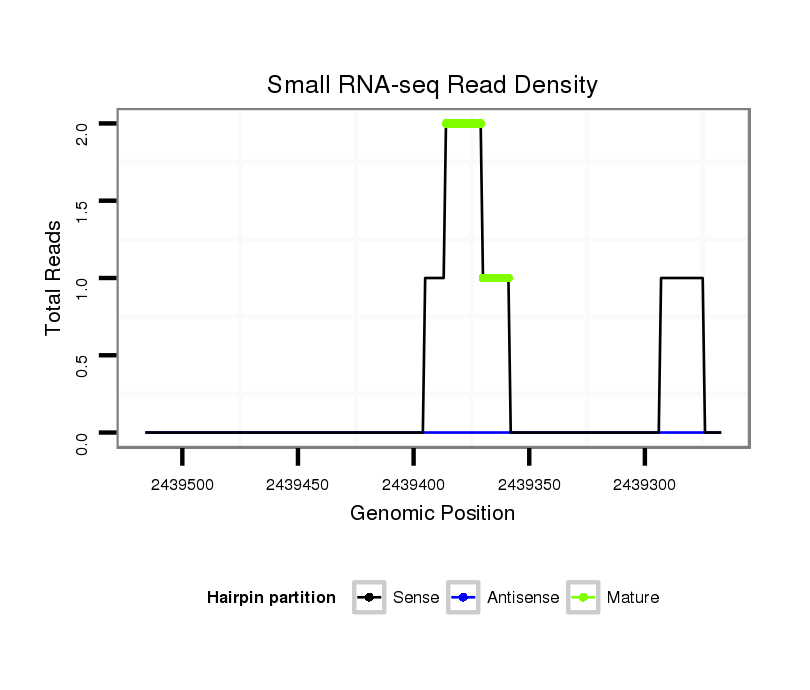
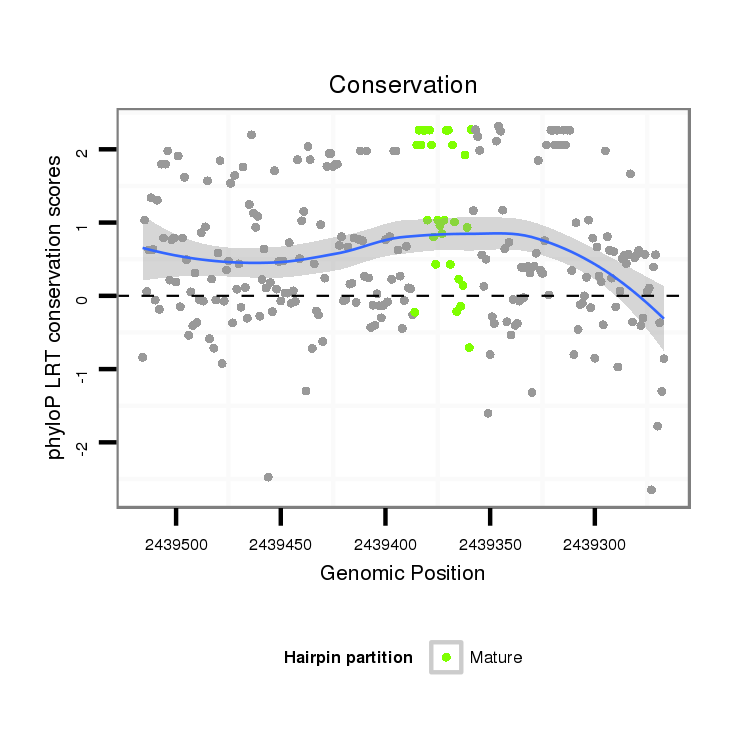
| droSim2 |
x:2439267-2439516 - |
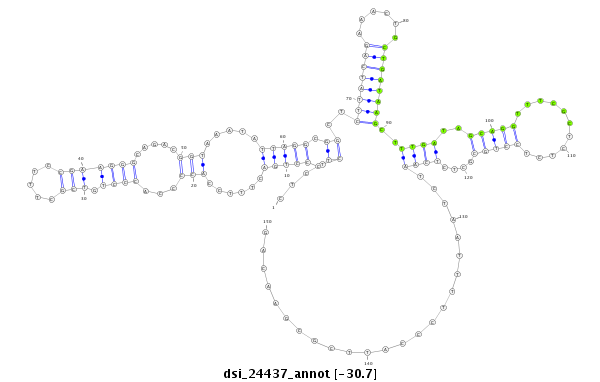
dsi_24437 |
G-----GCAAA-TA----------------------------------------TGTGATAG------GTGATAATCTGTGTTT---AGGCCT------------CCCTCT----------------------------------------------------CTCGCTCTCTCTCC--CTGACTT-TCCACCCCACCCTGTCGCT--TTC--CGAAGGGCAGAC---------------------------------GGTAAATATT---AGGC---GGCTCTTATCAGAAAC--T-----GCTGATAAGCTTT---GATAGCAG-----------------------------------------------------------------GTTT--------------------C-------------------------GCTCTC--TCC----T--GCGCT------CTCAA--------------------------------TCT-AATTTTCCCA----------------------TT-CGCGAACAGCTGTTGGAGG-CAAA--------------------------CGGC---AAC---------------AACAA---A-----------------------------------------------CCAGGAAC-AAAGCTCAC--AAGCCG-- |
| droSec2 |
scaffold_10:2398823-2399071 - |
|
G-----GCAAA-TA----------------------------------------TGTGATAG------GTGATAATCTGTGTTT---AGGCCT------------CCCTCT----------------------------------------------------CTCGCTCTCTCTCC--CTGACTT-TCCACCCCACCCTTTCGCT--TTC--CGAAGGGCAGAC---------------------------------GGTAAATATT---AGGC---GGCTCTTATCAGAAAC--T-----GCTGATAAGCTTT---GATAGCAG-----------------------------------------------------------------GTTT--------------------C-------------------------GCTCTC--TCC----T--GCGCT------CTCAA--------------------------------ACT-AATTTTCCCA----------------------TT-CGCGAACAGCTGTTGGAGG-C-AA--------------------------CGGC---AAC---------------AACAA---A-----------------------------------------------CCAGGAAC-AAAGCTCAC--AAGCCG-- |
| dm3 |
chrX:2637631-2637900 - |
|
G-----GCAAA-TA----------------------------------------TGTGATAG------GTGATAATCTGTGTTT---AGGCCTTTCTGACTG----------TCC----CTCTCTCTCTCTC----------------------------------TCTCT--CTCTCCATGACTT-TCCACCCCGCCCTGTCGCT--TTC--CGAAGGGCAGAC---------------------------------GGTAAATATT---AGGC---GGCTCTTATCAGAAAC--TTGTGCGCTGATAAGCTTT---GATAGCAG-----------------------------------------------------------------GTTT--------------------C-------------------------GCTCTC--TCC----T--GCGCT------CTCAA--------------------------------TCA-AATTTTCCCA----------------------TT-CGCGAACAGCTGTTGGAGG-C-AA--------------------------CGGC---AAC---------------AACAA---A-----------------------------------------------CCAGGAAC-AAAGCTCAG--AAGCTG-- |
| droEre2 |
scaffold_4690:63954-64194 - |
|
G-----GCAAA-TA---------------------------A--TATAAGTATATGTGATAG------GTGATAATCT-------------CTCTCT--------G------------------------------------------------------------TCTCTCTTTTG--CTGAATT-TCCACCTCGCCCTTTC---------------GGTAGGC---------------------------------GGTAAATATTT--AGGC---GGCTCTTATCAAAAAC--TTGCGCGCTGATAAGCGTT---GATAGCAA-----------------------------------------------------------------ACTT--------------------C-------------------------GCTCTC--TTC----T--GCGCT------CTCAA--------------------------------ACT-AATTTCCACA----------------------TT-CGCGAACAGCTGTTGGAGG-C-AA--------------------------CGGC---AAC---------------AACAA---G-----------------------------------------------CTCAGCAC-AAAGCTCGA--CAACCG-- |
| droYak3 |
X:5712324-5712571 + |
|
G-----GCAAA-TA----------------------------------------TGTGATAG------GTGATAATCT-------------------------------------------------------------------------------------CCCGCTCTCTCTCT--CTAAATT-TCCACCCCGCCCTGTCGCT--TTT--CGAAGGGCAAGC---------------------------------GGTAAATATTT--AGGC---GGCTCTTATCAAAAAC--TTGCGCGCTGATAAGCTTT---GATAGCAG-----------------------------------------------------------------GTTT--------------------C-------------------------GCTCTC--TTC----C--GCGCT------CTCAA--------------------------------TCT-AATTTTCCCA----------------------TT-CGCGAACAGCTGTTGGAGG-C-AA--------------------------CGAC---AAC---------------AACAA---A------------------CTCAGAAC------------------CAACTCAGAAC-AAAGCTCAG--AACCTG-- |
| droEug1 |
scf7180000409061:156960-157275 - |
|
G-----GCAAA-AA----------------------------------------TGTGATAA------GTGATACTATATGTTT---AGCCCTCT----C------T----TTGT----CTTCTG-TATACCCTTCTCTATCTTTCATTTGCCACCCCTCACTCTCTCTCTCT-------------------CTCTCTCTCTCTTTTATTG--TGAAGGGCATTC---------------------------------CGTAAATATTT--AGGC---GGCTTCTATCAAAAAC--CCGCCAGCTGATAAGCTTT---GATAACAGATAATGTGA-CAAG----------------------------------------------------CTT--------------------C-------------------------GCTCTCTCCTC----G--GTGCT------CTCAA--------------------------------TCT-TAATTTCTCA----------------------TT-CGCGAACAGCTGTTGAAGG-C-AA--------------------------CAACAACAAT---AACAAAGACAACAACT------------------------------------------------TAAAGTAAGAAC-AGAGCTCAA---AGCTG-- |
| droBia1 |
scf7180000302126:2707787-2708062 + |
|
G-----GAAAA-AA----------------------------------------GGTGATAATA--TCGTAATACTCGC---------------------TGTCTCTCTCCTCCGCCG--------TATACC------------------------------------CCTCTCTTT--CCAATTT-GCCACCCCTCTCTTTCGCC--GTG--TGAAGGGCATTC---------------------------------CGTGAATATTT--GGGC---GGCTCTTATCAGAGGC---CGCCAGCTGATAAGCTCT---GTTAACAGATAAGGTGA-CAAG----------------------------------------------------TTT--------------------C-------------------------CCTCTC--TTC----G--GCGCT------CTCAA--------------------------------TCTTAGTTTTCTCA----------------------TT-CGCGAACAGCTGTTGAAAG-C-CT--------------------------TAGC---TAC---------------AACAA---A---------------------------------------------AATTAAGAAAAAAGAATTAA--AAATAA-- |
| droTak1 |
scf7180000415769:1011017-1011290 - |
|
G-----GCAAA-AA----------------------------------------GGTGATAA------GTGATACCCGC---------------------TGTCTCTCTCCATCGCCA----------TATTCC----------------------------------TGTCTCTTT--CCCATTT-TCCACCCCTCTCTTTCGCC--TTG--TGAAGGGCATTC---------------------------------CGTAAATATTT--AGGC---GTTTCTTATCGGAAGC--CCGCCAGCTGATAAGCTTT---GATAACAGACAATGCGA-CAAG----------------------------------------------------TTT--------------------C-------------------------ACTCTC--TTC----G--CCGCT------CTCAA--------------------------------TCTTAATTTTCCCA----------------------TT-CGCGAACAGCTGTTAAAGG-C-AA--------------------------CAAC---AAC---------------AGCAA---A---------------------------------------------AATTAAGAAC-AAAGCTCAAACAGACCG-- |
| droEle1 |
scf7180000490141:342582-342854 + |
|
G-----GCAAA-AA----------------------------------------GGTGATAA------GTGATACTAGGGGCTT---TGCTCTCT----C------T----ATCA----CTCTCTCAATAACCC------------------------------------TCCCTCT--CTTATTTGTTCTCCCCTCTCTTTCTCA--TTG--TGAAGGGCATTC---------------------------------CGTAAATATTT--AACC---GGTCATTATCAA--------TTCACCTGATAAGCTTT---GATAACAGATAAAGCGA-AAAG----------------------------------------------------TTT--------------------C-------------------------GCTCTC--TTT----T--GCGCT------CTTCA--------------------------------TAT-AGTTTTCTCA----------------------TT-CGCGAACAGCTGTTGGAGC-C-AA--------------------------CAAC---AAC---------------AACGA---C---------------------------------------------AATTAAGAAC-AAAGCTCAA---AACCG-- |
| droRho1 |
scf7180000779467:378067-378337 + |
|
G-----GCAAA-AA----------------------------------------GGTGATAA------GTGATAGTAGGGGCTT---CGCTCTCT----C------T----ATCA----CTCTTACAATACCCC------------------------------------TCCCACT--CTAGTTTGTTCACCCCTCTCTTTCTCT--TTG--TGAAGGGCATTC---------------------------------CGTAAATATTT--AGGT---AGTCCTTATCAG--------GCCAGCTGATAAGCTTT---GATAACAGATAAAGTGA-CAAG----------------------------------------------------TCT--------------------C-------------------------GCTCTC--TTT----T--TCACT------CTCAA--------------------------------TTCTAGTATTTCCA----------------------TT-CGCGAACAGCTGTTGAAGG-C-AA--------------------------CAAC---AAC---------------AA------A---------------------------------------------AAATAAGAAC-AAAGCTCAG---AACCG-- |
| droFic1 |
scf7180000454072:601126-601417 + |
|
G-----GCAAA-AA----------------------------------------GGTGATAATACATAGTGA-----TGTGTTT---TGTTCTCT----C------T----AACGTGGTCTATTGAAAAGCCC------------------------------CTCTCTTTCTCTCT--CTAATTT--GCAATCGTCTTTTTCTCT--TTGTGAGAAGGGCTTTC---------------------------------CGTAAATGTAT--ATGCAAGGTTTCTTATCAGAAAC------GCGCTGATTAGCTTT---GATAGCAGATAGAGTGC-TAAA----------------------------------------------------TTT--------------------C-------------------------CCTCTC--TTT----T--GCGCT------CTCGA--------------------------------GCT-CATATTCTCA----------------------TT-CGCAAACAGCTGTTGGAAG-T-AA--------------------------CAGC---AAC---------------AACAA---A---------------------------------------------AATTAAGAAC-AAAGCTCGA--AAGCCAGA |
| droKik1 |
scf7180000302592:1968116-1968453 - |
|
T-----GCAACTAA----------------------------------------GGTGATCA------GTGATAACTT-------------CTCTCC--------G----------------------------------------------------CCGCTCTCTCCGGCTCTC---------------------TCG---CTC--TTT--TGAAGGGCATTCCC-------------------------------CGTGAATTTTTTTCGGC---TGCTCTTATCGAAAAC--TCGCTAGCTGATAAGGTTT---GATAACAGATAAGGTGGCGGTTCTCTGTTCTCACCTCTCTCGTCTCTGCTCCCATTGCT---------------------------------------------------CACCTCCTCTC--TCTC--TCTCTCTCGCTTTTCCTTTCGCGCG-----------------------TCA--------ATCG-TATTCTCTCATTGTCCGTTAAA-----------T-CGCGAACAGCTGTTGAAAG-C-AAGTGGCT------------------GCAAACAAAGCT---CAAGAGTGCAGCAACAA---A---------------------------------------------A-ACAAAAAG-GAAATTCAC--AAGTCG-- |
| droAna3 |
scaffold_13248:434288-434616 + |
|
T-----GAAAA-AATA-------------------------G------------TGTGAAAA------T-TATTATTT-------------CTTTCT--------CGGTCT----------------------------------------------------CGGACTATCTCTT---------------------TCT---GCC--AAG--TGAAGGGCACATATTTTGGAATA----TATATATACCTCTATATATATGTACATAT--ATAT---ATTTCTAATCAGCAAACGTCGTAAACTGATAAGCTTTTTTGATAACAT-----------------------------------------------------------------GCCTTAGTTCTCTTTGCACTTT-TCTCG-------------------CTCTCTCTC--TCT----G--ATTCT------CTCGAATTCTCGC--ATTCTCC---------------------------GGTTGTACGTAAGA-----------A-AGCAAACAGCTGTTAGGTT-A-AA--------------------------AAAT---ATAAATAAT---------AATAATAAAAGTAATATATATATTATATTTATATAT-------------------------------ATATC-C--AAGTTC-- |
| droBip1 |
scf7180000394734:47587-47914 - |
|
A-----GAAAA-TA---------------------------G------------TGTGA-AA------ATGATTATTT-------------CTTTCT--------G------------------------------------------------------------ACTATCTCTC---------------------TCT---GCC--AAG--TGAAGGGCACATATTTTGGAATACTCATATATATACTTCTATATCTGTATATATAT--TTTT---GTTTCTAATCAGTAAACGTCGTAAACTGATAAGCTTTTTGTATAACAA-----------------------------------------------------------------GCCTTAGTTCTCTTTGCACTTT-TCTCA-------------------C--TCTCTC--CCC----G--ATTCT------CTCAAATTCTCGC--ATTCTCC---------------------------GGTTGTACGTAAGA-----------A-AGCGAACAGCTGTTAGGTTGA-AA--------------------------AAAT---AAT---------------AATAT---A----------------TATTCATATATATATCCATGTTCCAGTTAAGTTAAGAAATAAAACATAT--A-AATA-- |
| dp5 |
XL_group1e:2031604-2031920 - |
|
AAAATTGCAAC-AA----------------------------------------AGAGACAA------GTTT----CT-----------------G----TGGCTCCCGCT----------------------------------CGCTTG-------CC--------GCTCTCTCT--CAGA---------CCCTCTCTATCGCT--CTCTGTGAAGGGCATTC---------------------------------CACAAATATTT--CGTT-TTGTTTTTGATAAGCATT--CAGCCGCCTGATAAGCTTC---GATAACAGTTAAACGGC-ACAGCTTCGCGCGCG----------------------------------------------------------CTCTTTCTCTCTCCATCACCACCTGCTCTC--ACGC--GCTCTTTT--------------------GCTCTCGCGTACCTGC-GGGATA--------GG--------------------CAGGAACACGCACAGTC-CCCGAACAGCTGTTGGAAC-G--AGAGTCATTCAATGACCAGCCGGTTG-GAGC---AAC---------------AACAA---G--------------------------------------------------------------------------- |
| droPer2 |
scaffold_18:882010-882331 - |
|
AAAATTGCAAC-AA----------------------------------------AGAGACAA------GTGT----CT-----------------C----TGGCTCCCGCT----------------------------------CGCTTG-------CC--------GCTCTCTGT--CAGA---------CCCTCTCTATCGCT--CTCTGTGAAGGGCATTC---------------------------------CACAAATATTT--CGTT-TTGTTTTTGATAAGCATT--CAGCCGCCTGATAAGCTTT---GATAACAGTTAAACGGC-AAAGCTTCGCGCGCG----------------------------------------------------------CTCTTTCTCTCTCCATCACCACCTGCTCTC--ACGC--GCTATTTT--------------------GCTCTCCCGTACCTGC-GGGATA--------GG--------------------CAGGAACACGCACAGTC-CCCGAACAGCTGTTGGAAC-G--AGTGTCATTCAATGAGCAGCCGGTGG-GAGC---AAC---------------AACAAGAAA-----------------------------------------------A------------------------C-- |
| droWil2 |
scf2_1100000004909:1948983-1949158 - |
|
C-----AATAA-CC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTGATAAGCATT---GATAACGAA----------AAGCTTC-----------TCTCTCTTCAACTCTTCTTGGTGAAGTCGAACACGGGTTT--------------------T-------------------------TCTCTC--TCT----G--ACGCT------CTCGTTGTCTCAC--ATCTGCTGGGAGAGGAAAGCAAG--------------------CAGGAACACGCAGAGTTTTGCAAACAGCTGTTGCAGC-C-CAGTG------------------------------------------------------------------------------------------------------------------------------TCAG-- |
| droVir3 |
scaffold_12970:3279131-3279193 - |
|
T---------------------------------------------------------------------------CT-----------------C----TGTCTCTCTCTCTCTCTC----------TCTCTC----------------------------------TCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------C-------------------------TCTCTCTCTCTCTCTCTCTGTCTGTCTCTCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:22358671-22358749 + |
|
ATAAA---CGA-TA---------------------------AGCGATAAGCATA-----------------------------TAGAACGC--------------G----------------------------------------------------CCGCTCTCTCTCTCTCTCT--CTGTCTA-TCTCTCTCCCCCTGTCTCT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15110:5818570-5818674 - |
|
G-----GCCAA-AATGTGGCTAAAAATAGCAAAGCGCAGACA--GTAAAGTGTT-----------------------------TGCGACGCTT------------CTCTCG----------------------------------------------------CTCTCTCTCTCTCT--CTCACTC-TCTCTCTGTCTCTGTCTCT--GTC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |