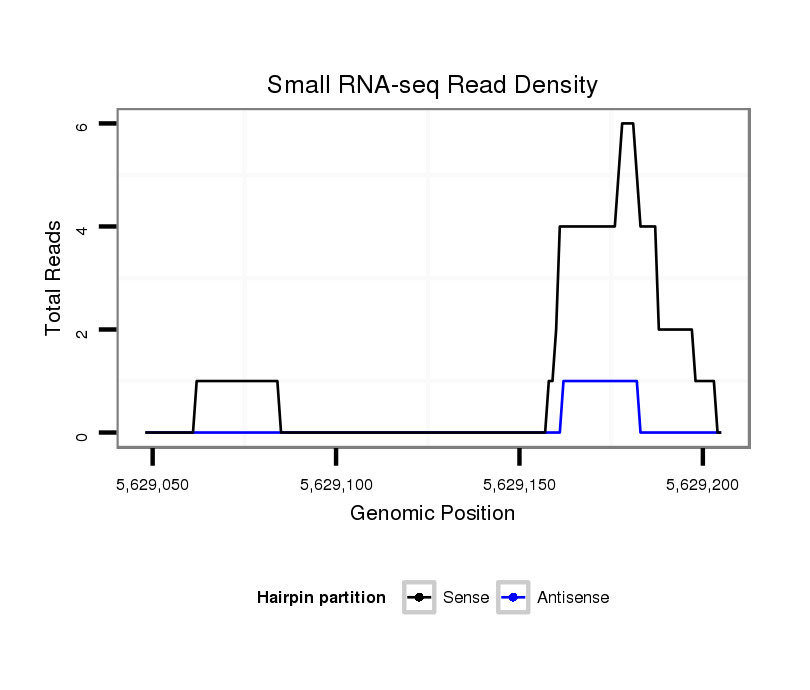
ID:dsi_24436 |
Coordinate:2r:5629098-5629155 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5628876_5629097_+]; CDS [2r_5628876_5629097_+]; CDS [2r_5629156_5629265_+]; exon [2r_5629156_5629265_+]; intron [2r_5629098_5629155_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------################################################## GTTGACGTGGGAGTAATATTTGTCGGACATGGTCTGAAGAACGACTTCAGGTGGGATGCCCTGCTTTATAGAGAACCGACAGACCACTAAACTTTCATGTGATTGCAGGGTCATAAACATCTACGTACCGTCGGAGCAGATAATAGACACGGTGCACC **************************************************((((..((.....(((....)))......))..)))).....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M053 female body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................TAAACATCTACGTACCGTCGGAGCAGA.................. | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CATCTACGTACCGTCGGG....................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GTCGGAGCAGATAATAGACAC........ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TCGGAGCAGATAATAGACACGGTGCA.. | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............AATATTTGTCGGACATGGTCTGA......................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................ATAAACATCTACGTACCGTCGGA....................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TCGGACATGGTCTGAAGAACGACTTTAGGGT......................................................................................................... | 31 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TCATAAACATCTACGTACCGTCGG........................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................TGGTCGGAAGAACGA.................................................................................................................. | 15 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..TGACGTGGGAGAAATT............................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................GTATCGTCGAAGCAGA.................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CAACTGCACCCTCATTATAAACAGCCTGTACCAGACTTCTTGCTGAAGTCCACCCTACGGGACGAAATATCTCTTGGCTGTCTGGTGATTTGAAAGTACACTAACGTCCCAGTATTTGTAGATGCATGGCAGCCTCGTCTATTATCTGTGCCACGTGG
**************************************************((((..((.....(((....)))......))..)))).....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TTTGTAGATGCATGGCAGCCT....................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CATGGCAGACTCGTCTATTAT............. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................TCTTGGTTGACTGGTGAT..................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................ACACCCTGTACCAGCCT......................................................................................................................... | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................TCGTGGCAGTCTAGTGAT..................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................CTCTTGGCTGGCAGGT........................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 08:08 PM