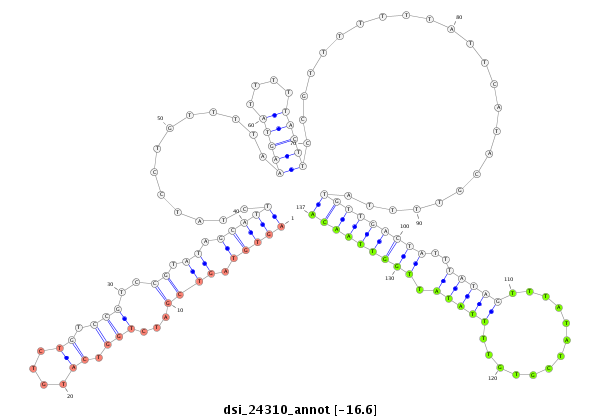
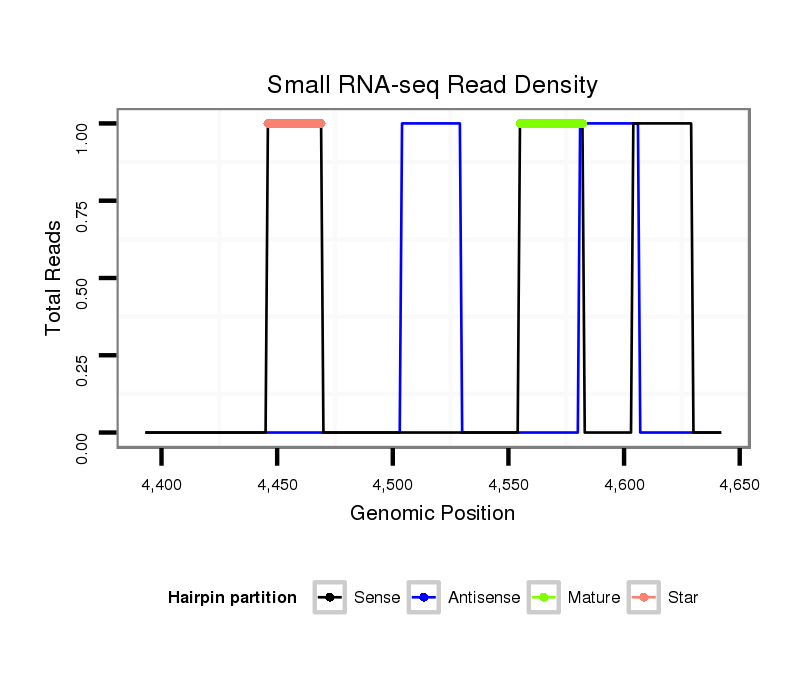
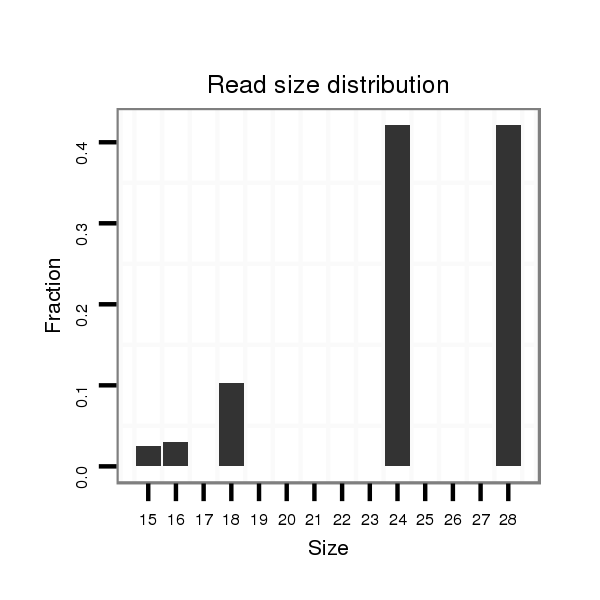
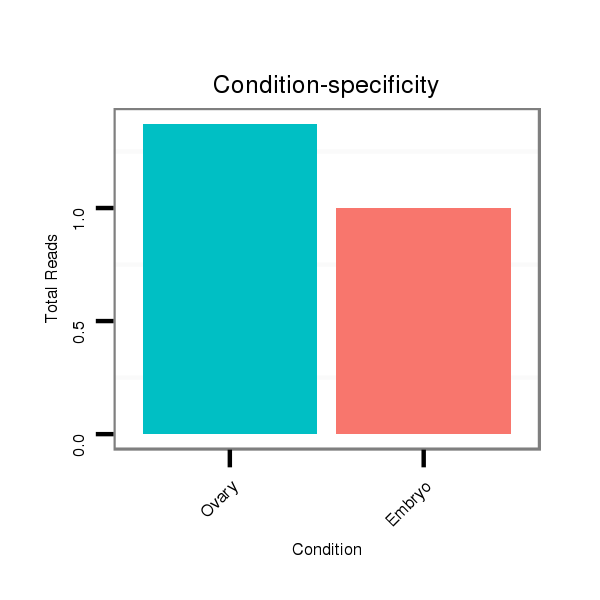
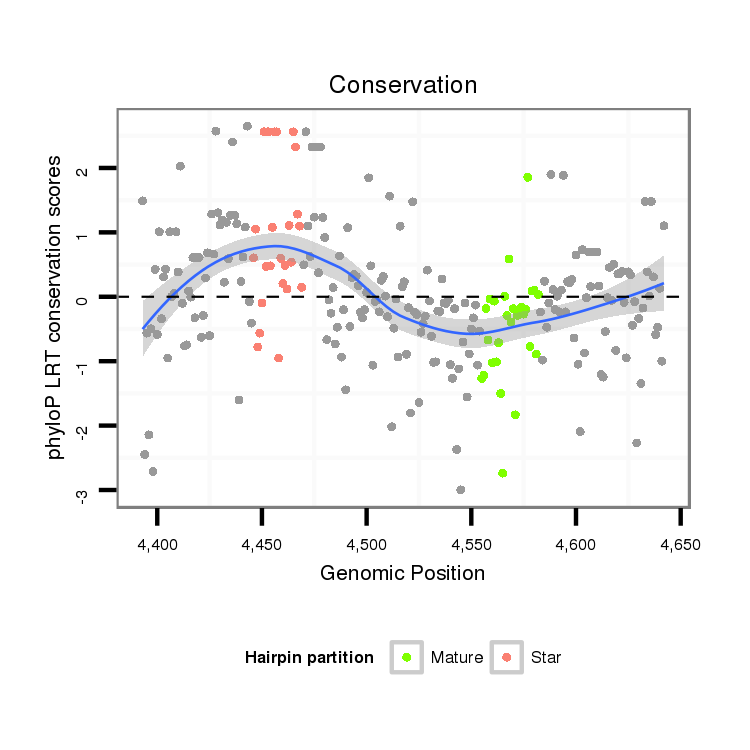
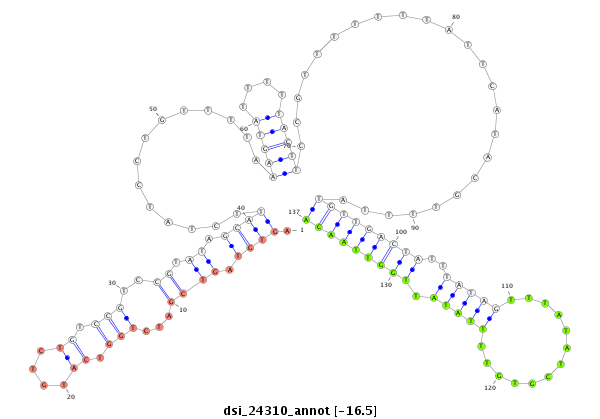
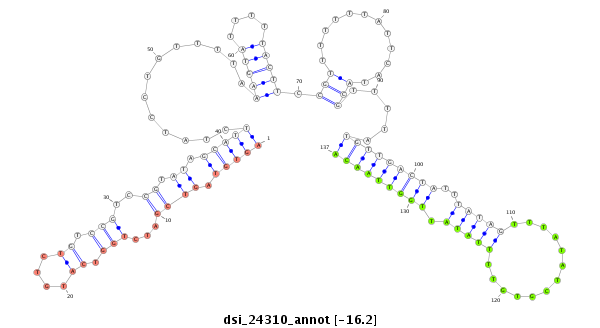
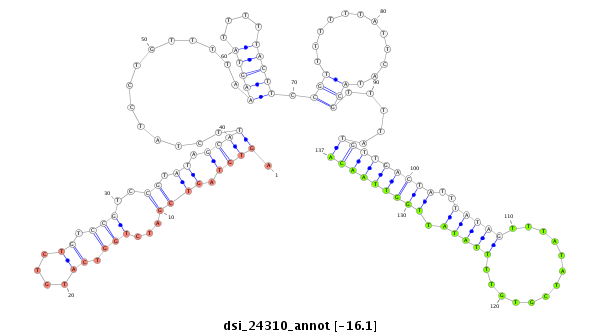
| TTGTGGTGAAAATGGATGTGTGTAACGTCCAGAAGGAATCGTTTCCGACCCCATAAAGTATATA-----------------TA-------TTC----TTGATCAGCATCAATAGCCAAGTCGATTGAGCCATGTCTATCTGTCCGTCTGTCCGTCCGTCC-----------GT--C--TGTCCGTCC--------------------------GT-----------CT----GTCTGTCCGTCCCTATTA------------------------------------GCGCCGAGTGCT--------------------CA------------------AAGACT---------------------------------------------------------------------------------------------------------------------------------------------------------------------ATA | Size | Hit Count | Total Norm | Total | M021
Embryo | M042
Female-body | V042
Embryo | V050
Head | V057
Head | V111
Male-body |
| .........................CGTCCAGAAGGAATCGTTTC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 1.80 | 36 | 0 | 0 | 0 | 0 | 0 | 36 |
| ................TGTGTGTAACGTCCAGAAGGAATCGTT..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.50 | 10 | 0 | 10 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TGAGCCATGTCTATCTGTCCGTCTG........................................................................................................................................................................................................................................................................................................................................................................... | 25 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......GAAAATGGATGTGTGTAACG..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.25 | 5 | 0 | 0 | 0 | 0 | 0 | 5 |
| ...........................................................................................TC----TTGATCAGCATCAATAGCCAAGTCGA..................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| ...........................................................................................TC----TTGATCAGCATCAATAGCCAAG......................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGATCAGCATCAATAGCCAAGTC....................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 | 0 |
| .........AAATGGATGTGTGTAACGTC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................C-----------GT--C--TGTCCGTCC--------------------------GT-----------CT----GTC..................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TCAGCATCAATAGCCAAGTCG...................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................CC--------------------------GT-----------CT----GTCTGTCCGTCCC........................................................................................................................................................................................................................................................................... | 19 | 20 | 0.10 | 2 | 0 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................CAAGTCGATTGAGCCATGTCT........................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................TC----TTGATCAGCATCAATAGCCAAGTCG...................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................CATCAATAGCCAAGTCGATTG.................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAGTCGATTGAGCCATGTCT........................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGATCAGCATCAATAGCCAAG......................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAGTCGATTGAGCCATGTC......................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TCCGTCC-----------GT--C--TGTCCGTCC--------------------------GT-----------CT----GT...................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................GCATCAATAGCCAAGTCGATT................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGATCAGCATCAATAGCCAAGTCGAT.................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................ATCAATAGCCAAGTCGATTGA................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................C----TTGATCAGCATCAATAGCCAAG......................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................AGCATCAATAGCCAAGTCGAT.................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................ATCAGCATCAATAGCCAAGTC....................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TC----TTGATCAGCATCAATAGCCAAGT........................................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................GCCAAGTCGATTGAGCCATGT.......................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGATCAGCATCAATAGCCAAGTCGATT................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................TTGATCAGCATCAATAGCCAAGT........................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |