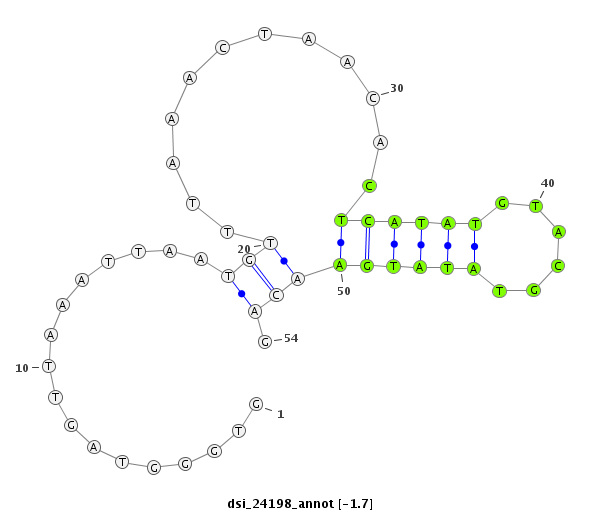
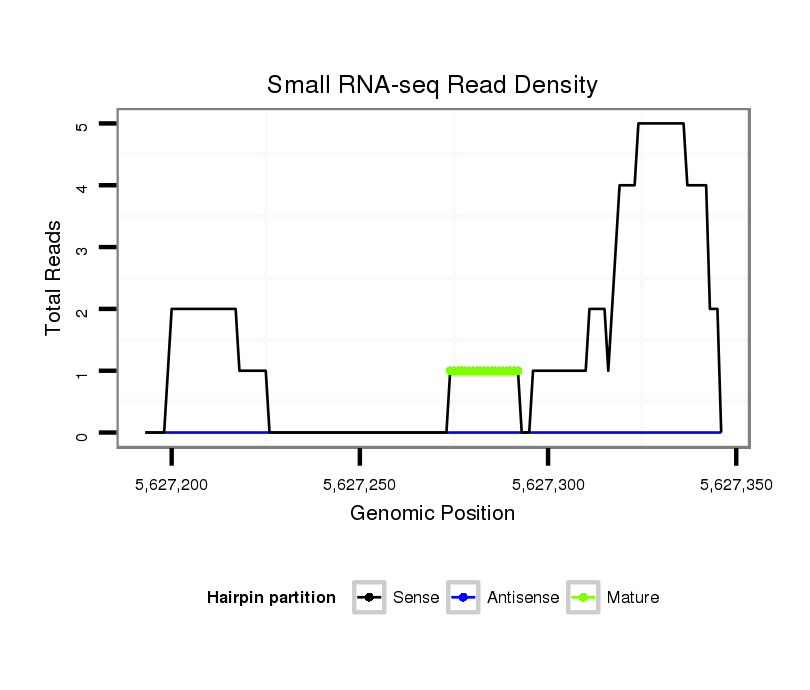
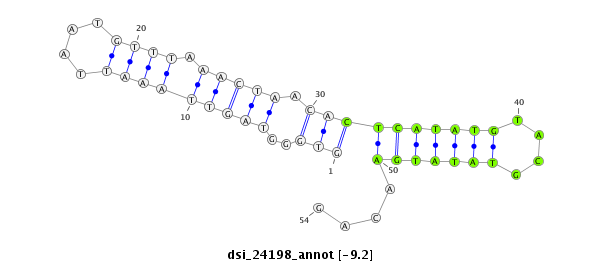
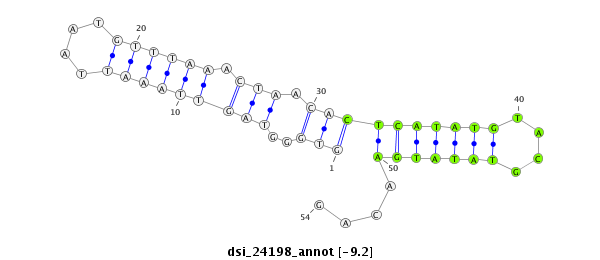
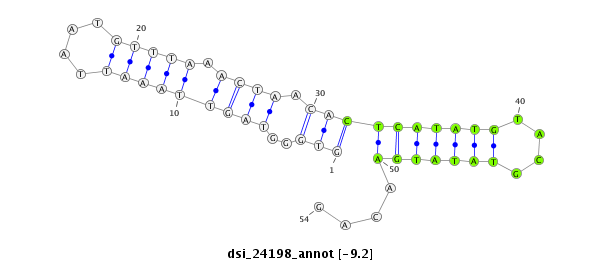
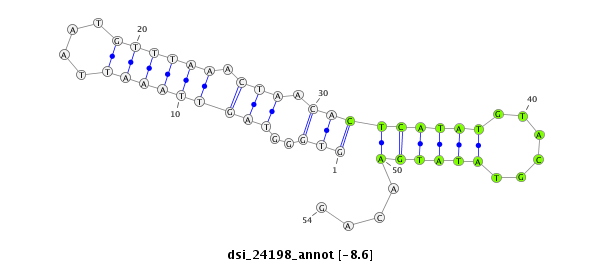
ID:dsi_24198 |
Coordinate:2r:5627243-5627296 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -9.2 | -9.2 | -9.2 | -8.6 |
 |
 |
 |
 |
exon [2r_5627183_5627242_+]; CDS [2r_5627183_5627242_+]; CDS [2r_5627297_5627592_+]; exon [2r_5627297_5627592_+]; intron [2r_5627243_5627296_+]
No Repeatable elements found
| ##################################################------------------------------------------------------################################################## GGCCACGTCGGAAACAAGTGGCTCCTCGGACCTTTACGATTCCATAAGTGGTGGGTAGTTAAATTAATGTTTAAACTAACACTCATATGTACGTATATGAACAGATGAGAACACCAAGGAGGACGATCGGGAACGCAGTAAGATCAACGCAGAA **************************************************.................(((............((((((......))))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
M024 male body |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M023 head |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
M053 female body |
SRR902009 testis |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................CTCCTCGGACCTTGAAAATT................................................................................................................. | 20 | 3 | 9 | 25.78 | 232 | 168 | 5 | 20 | 3 | 2 | 13 | 7 | 0 | 13 | 1 | 0 | 0 |
| .....................CTCCTCGGACCTTGA...................................................................................................................... | 15 | 1 | 5 | 2.20 | 11 | 0 | 5 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| ....................................................................................................AGTGATGAGAACACCAAGGAGGACGA............................ | 26 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................TGGAACGCAGAAAGATCAACGAAGAA | 26 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CTCCTCGGACCTTTAAAATTC................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......TCGGAAACAAGTGGCTCC................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................ATCGGGAACGCAGTAAGATCAACGCAGA. | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................AACGCAGTAAGATCAACGC.... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GATGAGAACACCAAGGAGGA............................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................CTCATATGTACGTATATGA...................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......GTCGGAAACAAGTGGCTCCTCGGACCT......................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GATCGGGAACGCAGTAAGATCAACGC.... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................TCGGGAACGCAGTAAGATCAACGCAGA. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................GAGGACGATCGGGAACGCAGTAAGAT.......... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GACGGAAACAAGTGGCGGCTC............................................................................................................................... | 21 | 3 | 5 | 0.60 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................CTCCTCGGACCTTTGCG.................................................................................................................... | 17 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................TTTCGATTCCCTAAGTGG....................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......GACGGAAACAAGTGGCGGCT................................................................................................................................ | 20 | 3 | 14 | 0.21 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................CGATTCCGTAAGTAGTGG.................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................ACACCAAGGAGGAAGA............................ | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................ATTTGCTCCTCGGACCTT........................................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............AACAAATTTCTCCTCGGACC.......................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................TGTCTCCTCGGACCT......................................................................................................................... | 15 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................ATTTCGATTCCCTAAGTGG....................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................TCCTCGGACCATTATGA................................................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................ATTTTCTCCTCGGACCTTTA...................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................TGGTGGGTAGTTTAACT......................................................................................... | 17 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................AAAGTGGTGGGTAGT............................................................................................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GGTGGGTAGTTTAACT......................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................AGGGGATCTTCGGACCTT........................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................AACTCAATGTTTAAACT............................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............ACAATTTTCTCCTCGGACC.......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
CCGGTGCAGCCTTTGTTCACCGAGGAGCCTGGAAATGCTAAGGTATTCACCACCCATCAATTTAATTACAAATTTGATTGTGAGTATACATGCATATACTTGTCTACTCTTGTGGTTCCTCCTGCTAGCCCTTGCGTCATTCTAGTTGCGTCTT
**************************************************.................(((............((((((......))))))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|
| ................................AAATCCTAAGGTATTCCCTAC..................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ...................CCGAGGATGCTGGAAAT...................................................................................................................... | 17 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| ..................ACCGAGGAGACTGGAAT....................................................................................................................... | 17 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 |
Generated: 04/23/2015 at 08:00 PM