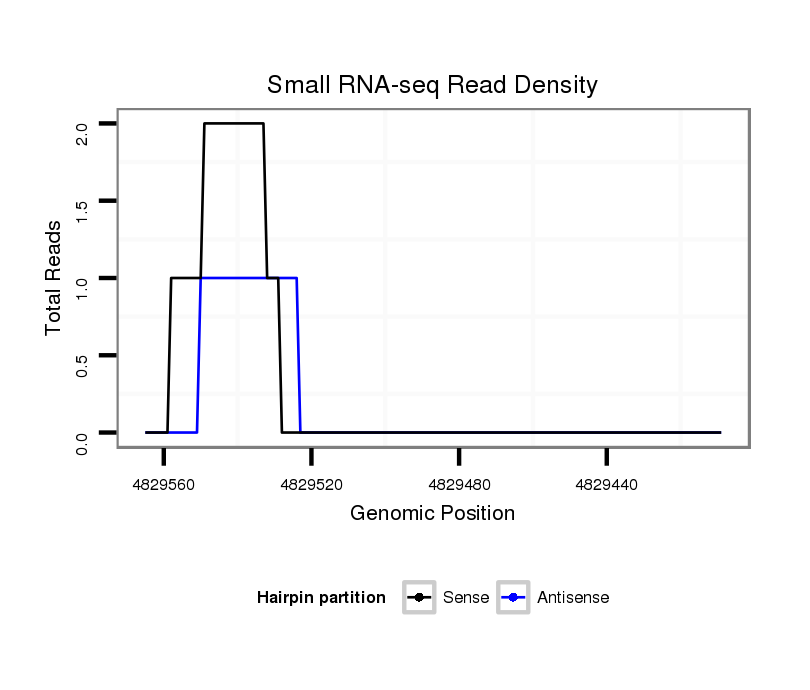
ID:dsi_23886 |
Coordinate:2r:4829459-4829515 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [2r_4829319_4829458_-]; exon [2r_4829230_4829458_-]; CDS [2r_4829516_4829722_-]; exon [2r_4829516_4829722_-]; intron [2r_4829459_4829515_-]
No Repeatable elements found
| ##################################################---------------------------------------------------------################################################## ATGCACTTACGTCAACCGGCGGACCAAAATTGTAACAGTGGAAACAATAAGTAAGCGACTATCAATTTAAATTGTATACCCGAAACTGACCACGTTAAAATTTGTAGTTTATCCCCCCGAAATCGGTCGCATATTGAGTCAATATTATATGAGCGGG **************************************************.....((..(((((((....)))).)))..)).........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR553488 RT_0-2 hours eggs |
M053 female body |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TATCACCCTGAAATCGGT.............................. | 18 | 2 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................TCGGAACTGACCACGGTAAAA......................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GTTTATCACCCTGAAATCG................................ | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTATCACCCTGAAATCGGT.............................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................CGGCGGACCAAAATTGTAACA........................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......TACGTCAACCGGCGGACCAAAATTGT............................................................................................................................ | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TTTATCACCCTGAAATCGG............................... | 19 | 2 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GTCTATCACCCTGAAATCGGT.............................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTATCACCCTGAAATCGG............................... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................AGCTTATCACCCTGAAATCGG............................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................TATCACCCTGAAATCGG............................... | 17 | 2 | 7 | 0.29 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................TGTAGACCCGAAACC...................................................................... | 15 | 2 | 20 | 0.25 | 5 | 1 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| ...........................................................................................................TCTATCACCCTGAAATCGGT.............................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................TATCCACCCGAAATCGG............................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTATCACCCCGGAATTGGT.............................. | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 |
| ...........................................................................................................TTTATCACCCTGAAATC................................. | 17 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TTAACCCCCTGAAATCGGTT............................. | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................ACCACGTTAATATTCGT.................................................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................TTGTAGACCCGAAACC...................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TACGTGAATGCAGTTGGCCGCCTGGTTTTAACATTGTCACCTTTGTTATTCATTCGCTGATAGTTAAATTTAACATATGGGCTTTGACTGGTGCAATTTTAAACATCAAATAGGGGGGCTTTAGCCAGCGTATAACTCAGTTATAATATACTCGCCC
**************************************************.....((..(((((((....)))).)))..)).........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902008 ovaries |
|---|---|---|---|---|---|---|
| ...............GGCCGCCTGGTTTTAACATTGTCACCT................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 |
Generated: 04/23/2015 at 06:33 AM