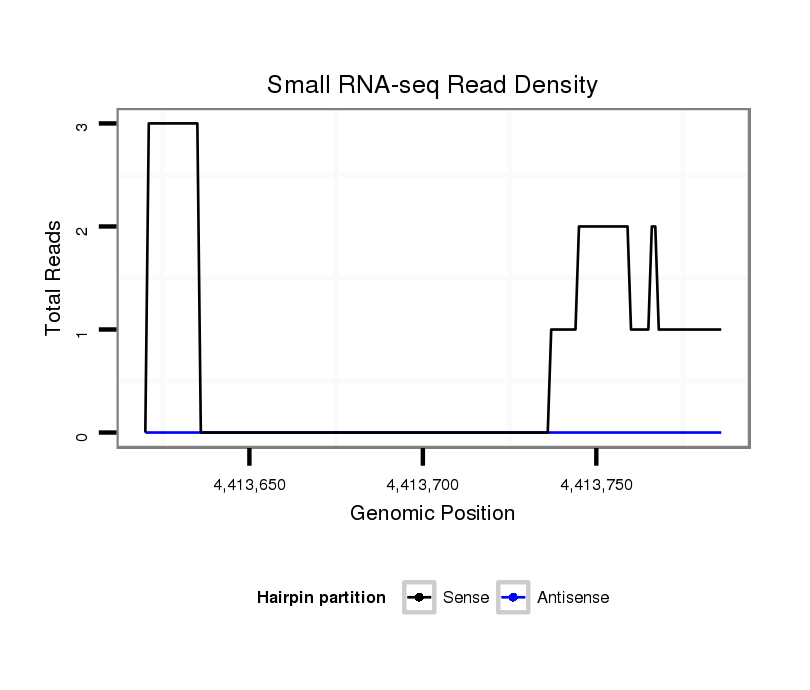
ID:dsi_23869 |
Coordinate:3r:4413670-4413736 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TGGGGATCGAAGACGATCGACGGTAGCGGTCGCACAGCAGGCACCATCGAGTGAGTCATCCGAAATTCTCGAAACTCTGGGTATCGGGTTCATTGTTCCCTTTCTTTCTGCTTTTAGCATGGACAAGCTGCGCGAGGAGACGGAGCAGGCCGATCTGGACAAGAAGA
**************************************************((((...((((((.((((..........)))).))))))))))........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
SRR618934 dsim w501 ovaries |
M023 head |
M053 female body |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................CAGTAGGCACCATCAA..................................................................................................................... | 16 | 2 | 20 | 140.20 | 2804 | 1850 | 531 | 423 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AGGGGATCGAAGATGATC..................................................................................................................................................... | 18 | 2 | 7 | 23.57 | 165 | 0 | 0 | 0 | 107 | 2 | 47 | 0 | 9 | 0 | 0 |
| ..............................CTCACAGGAGGCACCA......................................................................................................................... | 16 | 2 | 20 | 12.75 | 255 | 0 | 4 | 0 | 0 | 251 | 0 | 0 | 0 | 0 | 0 |
| .GGGGCTCGAAGACGATC..................................................................................................................................................... | 17 | 1 | 2 | 5.00 | 10 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGCTCGAAGACGAT...................................................................................................................................................... | 16 | 1 | 5 | 4.00 | 20 | 13 | 1 | 4 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| GGGGGCTCGAAGACGATC..................................................................................................................................................... | 18 | 2 | 2 | 3.50 | 7 | 3 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGCTCGAAGACGATCA.................................................................................................................................................... | 18 | 2 | 6 | 3.17 | 19 | 8 | 3 | 7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGACGA....................................................................................................................................................... | 15 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGATGATCA.................................................................................................................................................... | 18 | 2 | 3 | 3.00 | 9 | 1 | 0 | 1 | 4 | 0 | 2 | 1 | 0 | 0 | 0 |
| .GGGGCTCGAAGACGA....................................................................................................................................................... | 15 | 1 | 8 | 2.88 | 23 | 11 | 2 | 8 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| GGGGGCTCGAAGACGATCA.................................................................................................................................................... | 19 | 3 | 11 | 1.82 | 20 | 5 | 3 | 9 | 2 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................CGCACTGTAGGCACCATCAA..................................................................................................................... | 20 | 3 | 8 | 1.38 | 11 | 4 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| GGGGGCTCGAAGACGAT...................................................................................................................................................... | 17 | 2 | 12 | 1.25 | 15 | 12 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................AGCTGCGCGAGGAGACGGAGCAG................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| TGGGGATCGAAGATGATC..................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CATGGACAAGCTGCGCGAGGAGA........................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................TCGCACAGGAGGCACCA......................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGACGATCA.................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGGCCGATCTGGACAAGAAGA | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................TCGCACTGTAGGCACCATCAA..................................................................................................................... | 21 | 3 | 3 | 1.00 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................CGCACAGGAGGCACCA......................................................................................................................... | 16 | 1 | 4 | 0.50 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| AGGGGATCGAAGACGATCA.................................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGACGATCAGA.................................................................................................................................................. | 20 | 3 | 9 | 0.44 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGATGAT...................................................................................................................................................... | 16 | 1 | 5 | 0.40 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAGACTATCA.................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................GCCGATCTGGACAGGAA.. | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| GGGGGATCGAAGATGATC..................................................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| AGGGGATCGAAGACGA....................................................................................................................................................... | 16 | 1 | 7 | 0.29 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GGGCTCGAAGACGATC..................................................................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGCTCGAAGACGATCC.................................................................................................................................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................CAGCAGGCACCATCAA..................................................................................................................... | 16 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .GGGGATCGAAAACGATCA.................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| AGGGGATCGAAGACG........................................................................................................................................................ | 15 | 1 | 14 | 0.14 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........GACGATCGACGTCAGCTGT......................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................CTCGAAAGTATGGGTAGCG................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...GGCTCGAAGACGATC..................................................................................................................................................... | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................CGGACAGTAGGCACCATCAA..................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................AAGCTGCACGAGGAGA........................... | 16 | 1 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..GGGATCGAAGATGATCA.................................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AGGCCGTTATGGACAGGA... | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
ACCCCTAGCTTCTGCTAGCTGCCATCGCCAGCGTGTCGTCCGTGGTAGCTCACTCAGTAGGCTTTAAGAGCTTTGAGACCCATAGCCCAAGTAACAAGGGAAAGAAAGACGAAAATCGTACCTGTTCGACGCGCTCCTCTGCCTCGTCCGGCTAGACCTGTTCTTCT
**************************************************((((...((((((.((((..........)))).))))))))))........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................ACCCGTCCGGCTAGACCTG....... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................GGAAAGAAAGACGAA...................................................... | 15 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................GGAAAGAGAGACGAAAA.................................................... | 17 | 1 | 18 | 0.22 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| .................................................TCAGTCAATAGGCTTTAA.................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................ACTCGCTCCCCTGCCTCG..................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................GCTTTAAGGGCTTTGAG.......................................................................................... | 17 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AGAGCTTAGAGACCCA..................................................................................... | 16 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...CCAAGCTACTGCTAGCAGC................................................................................................................................................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 06:34 AM