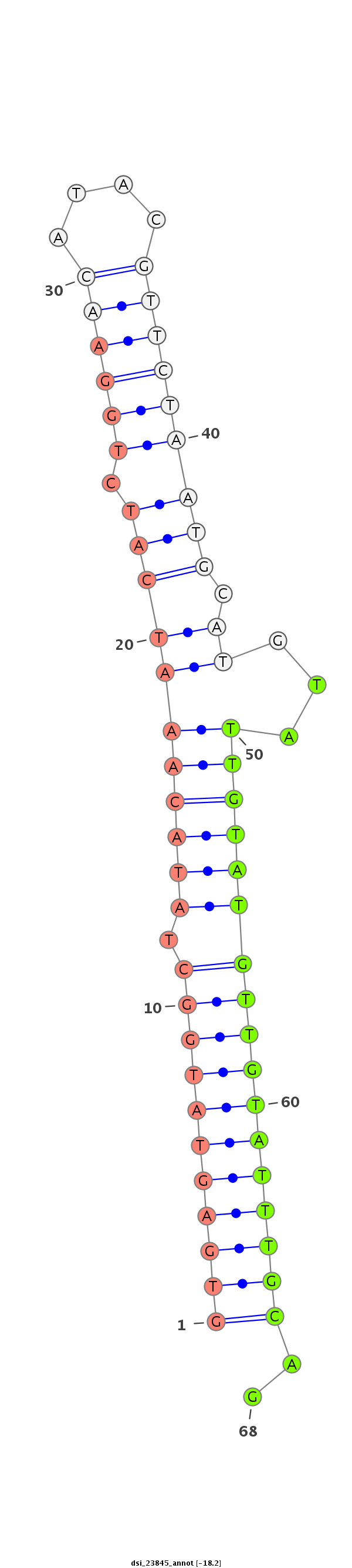
| droSim2 |
2l:13205271-13205438 + |
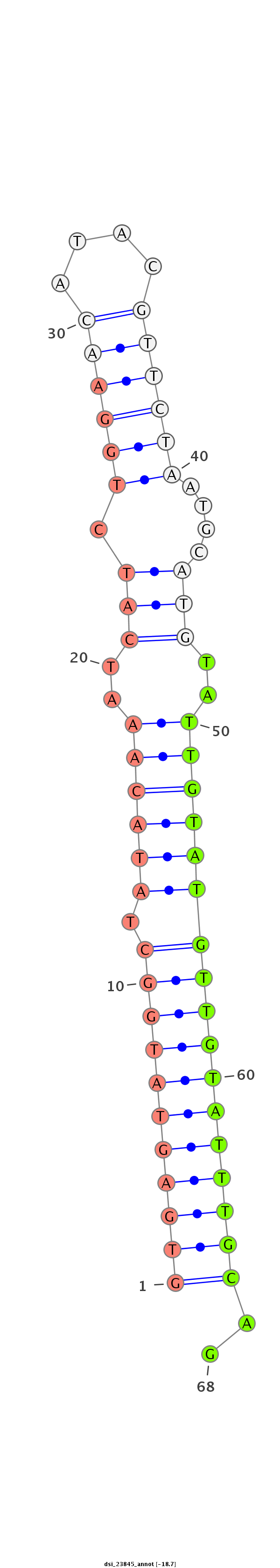
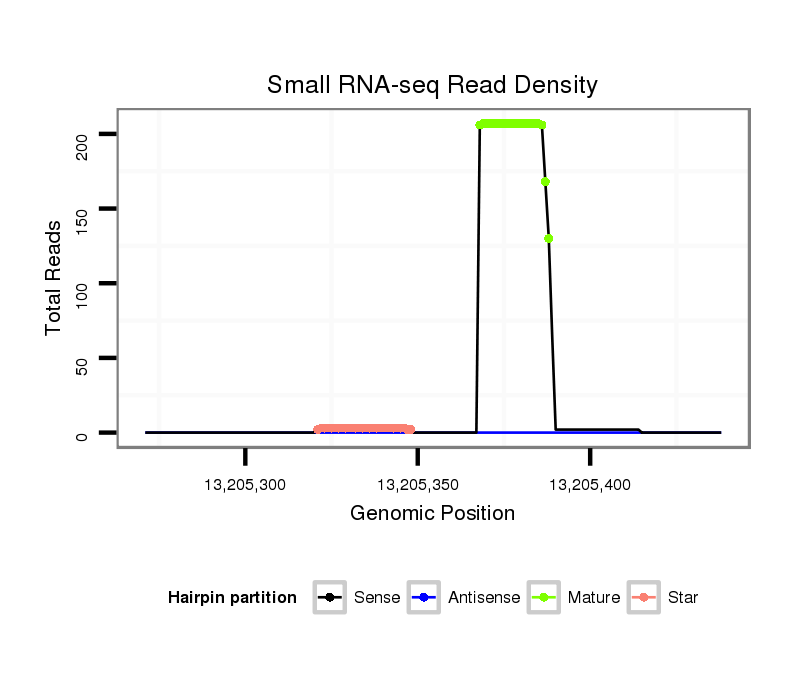
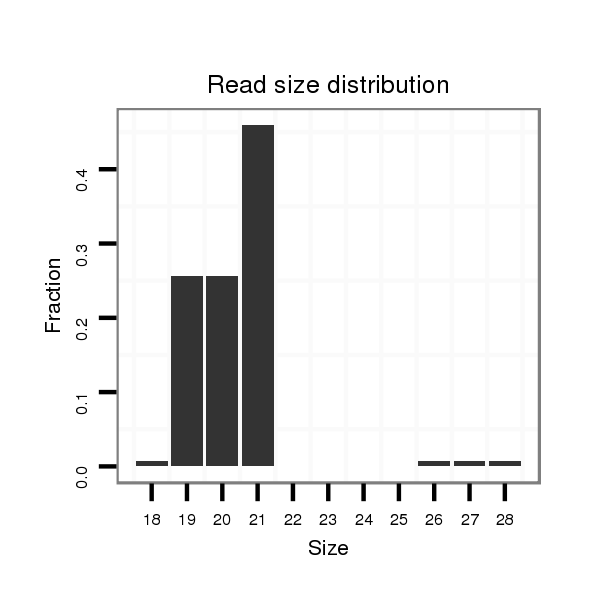
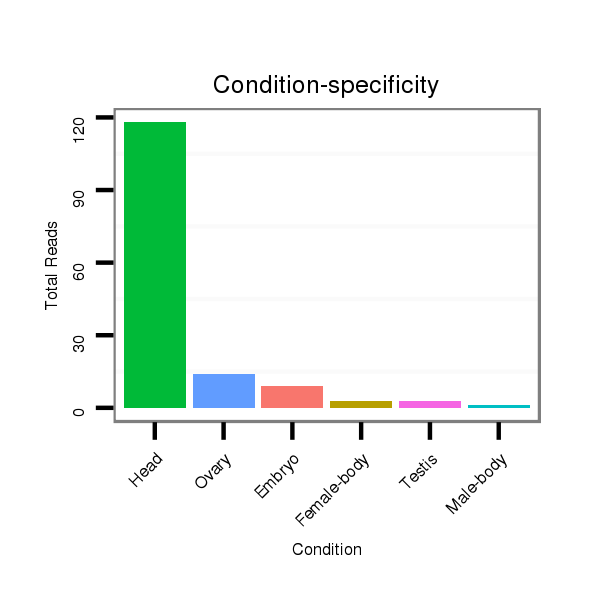
dsi_23845 |
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATGG------C--TATACA----AA-T--------------C-----------------ATCTGGAACATACGTTC-------TAAT-----GCATG----------------------T-----------------A-------------TTGTAT--GTTG-TATTTGCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droSec2 |
scaffold_16:1746015-1746182 + |
dse_353 |
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATGG------C--TATACA----AA-T--------------C-----------------ATCTGGAACATACGTTC-------TAAT-----GCATG----------------------T-----------------A-------------TTGTAT--GTTG-TATTTGCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| dm3 |
chr2L:13609938-13610105 + |
dme-mir-2489 |
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATGG------C--TATACA----AA-T--------------C-----------------ATCTGGAACGTAAGTTC-------TAAT-----GTATG----------------------T-----------------A-------------TTGTAT--GTTG-TATTTGCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droEre2 |
scaffold_4929:11847801-11847962 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATGG------C--TATCTA----AA-T--------------C-----------------AGCCG-AACGTGTATTC-------TAAT-----GT---------------------------------------------------------TTGCAC--TTTG-TATTTGCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droYak3 |
2L:10029671-10029836 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGATGTGAGTATGG------C--TATTTA----AA-T--------------C-----------------AACCG-AACGT------------------------ATGTATGT----------TATATAGT-------------------------------TTGCAT--TTTG-TATTTGCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droEug1 |
scf7180000409452:191269-191429 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATAG------T--ACT-TA----AA-T----TATA-------ATTTTAAAT------------------CACATTA-------TAA------------------------------------------------------------------TGTAC--TTTGTACTTTTCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droBia1 |
scf7180000302408:2742657-2742821 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGAGGTGAGTACTC------C--TA---A-----AATATACT---AAAA---ATAAGAAA--------------------ACATTC-------TAA---------------------------------C-----------------A-------------ACCAATA-TTTG-CCCTTGCAGATTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droTak1 |
scf7180000415285:144366-144531 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATAT------A--AATATA----TA-T--------------T-----------------AATTAAAATTTAAATCT-------TAAA-----TC--G----------------------T-----------------A-------------ACATTT--GTTG-TACTTTCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droEle1 |
scf7180000491338:1217874-1218038 + |
|
TCCATCTTAGACTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATTG------T--CATTAA---G-A-T----A---------------------------GAACA--ACAT------------------------AAACATGT----------TAAACA-T-----------------A----T--------TTGTAT--TTT--TTTTTTCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droRho1 |
scf7180000780107:475209-475370 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATTT------T--T---TA----AA-T--------------AATTTAAA---------------AAACTTACAACT-------TAAT-----GT--G----------------------T---------------------------------ACAC--CTTG-TATTTTCAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droFic1 |
scf7180000453925:389393-389555 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATTG------C--TCTCAA---G-A-T----TAAA-------ATCAAAAAC-------------------CCTTAC-------TAAG-----AC-----------------------A-T-----------------A-----------------CT--TTTT-TATTTTTAGTTTTATGATAGCAAAGGTCCCATTGATGTCTCCACGGATCATATCTATGA |
| droKik1 |
scf7180000302656:77420-77583 + |
|
TCCATCTTAGATTAAAGCGTGATTTAAATACATTTAGCAATAAGTTAGACGTGAGTATTAC-----A--TACG-C----AA-T--------------T-----------------ATATTAAT---------TTTAAGT-------------------AATGCTCG---------T-----------------A-------------TT-TCC--TTTA-CTTTTCCAGTTTTATGATAGCAAGGGCCCCATTGATGTCTCCACGGATCATATCTATGA |
| droAna3 |
scaffold_12943:1669964-1670138 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCGATAAGTTAGACGTGAGTATTGT-----C--TCTGTC----AA-G--------------T-----------------GTATGGTACTAATA---TTTAACT-------------------AATTCTTGTAT------T-----------------A-------------ACTTGT--TTTT-AATTTCCAGTTTTATGATAGCAATGGCCCCATTGATGTCTCCACGGATCATATCTATGA |
| droBip1 |
scf7180000396728:1558126-1558292 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCGATAAGTTAGACGTGAGTATTA------A--TATAAT---C-C-T----TAAA-------ATATAAAAT-------------------CAATAA-------TTAA-----TC--------A----------------A-----------------A-------------ACCTCT--TTCT-GATCCATAGTTTTATGATAGCAATGGCCCCATTGATGTCTCCACGGATCATATCTATGA |
| dp5 |
4_group1:1874114-1874281 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCGATAAGTTAGACGTGAGTATTT------T--CC---A-----TACCTTTT---TAAAACA-------ATCATTTTGA---TA-TACAT------------------------ATA----------------------T-----------------A-------------TTTTTT--GTTG-TG--TTTAGTTTTATGATAGCAACGGCCCCATTGATGTCTCCACGGATCATATCTATGC |
| droPer2 |
scaffold_5:3326139-3326312 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCGATAAGTTAGACGTGAGTATAT------C--TTTGCC----ATCTCTTTT---TAAAATA------------------------TTTTATATTT-------TTATATATTACATA----------------------T-----------------A-------------TTTTTT--G-----GTGTTTAGTTTTATGATAGCAACGGCCCCATTGATGTCTCCACGGATCATATCTATGC |
| droWil2 |
scf2_1100000004585:852516-852683 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGCGATAAGTTAGATGTGAGTATAT------T--AT----------------------------------------------------------------------------------------------------ATCTTTTGTCCCATTTTGGAATATGTTTTTGAATACCTTTTC--TTTG-GGATTTCAGTTTTATGATAGCAACGGCCCCATTGATGTCTCCACGGATCACATCTATGA |
| droVir3 |
scaffold_12723:5236320-5236483 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGTGATAAGTTAGACGTGAGTATTA------T--CA---ACATA-T-A----T---------------------------GAATT--ATT-------TTAAAAT-------------------AATAATCG---------A-----------------A--------------TTTAA--TTTG-TTATCCCAGTTTTATGATAGCAACGGCCCCATTGATGTCTCAACGGATCATATCTATGA |
| droMoj3 |
scaffold_6500:16444073-16444232 + |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGTGATAAGTTAGACGTGAGTATTACAGAGGCCCTATCAA----AA-T--------------A-----------------------------------------TCAT-----TCGTG----------------------T-----------------A-------------TCTAATATAAT--ATTTTTCAGTTTTATGATAACAACGGCCCCATTGATGTCTCAACGGATCATATCTATGA |
| droGri2 |
scaffold_15126:8263458-8263622 - |
|
TCCATCTTAGATTAAAACGTGATTTAAATACATTTAGTGATAAGTTAGACGTGAGTATTT------A--TT--AA------CTTCATT---CAAC------------------------GTAATATACATAC-------TAAA-----AT---------------------------------------------------------ATGAATACATC--ATTTTGCAGTCTTATGATAGCAACGGCCCCATTGATGTCTCAACGGATCATATCTATGA |