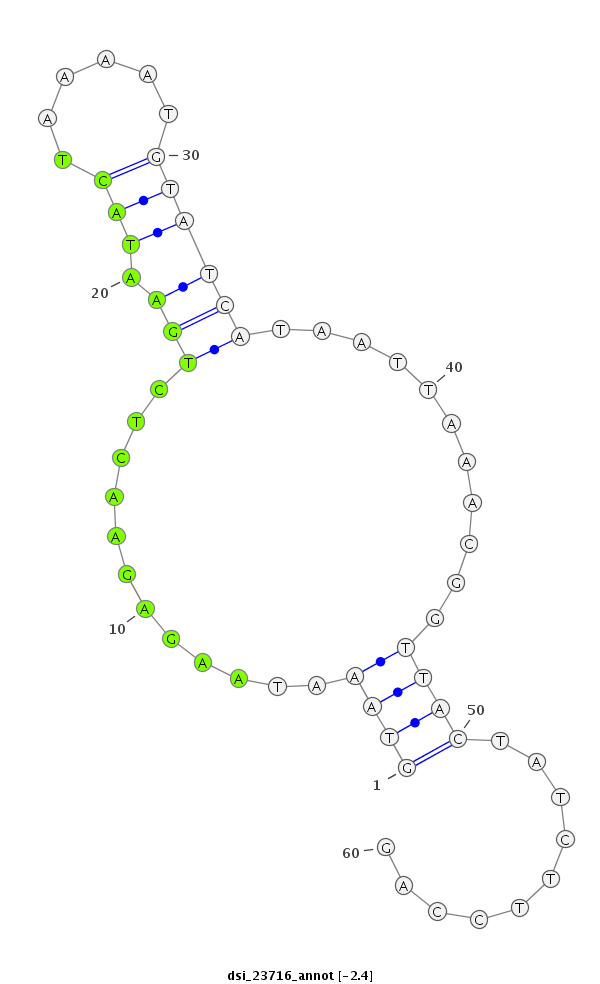
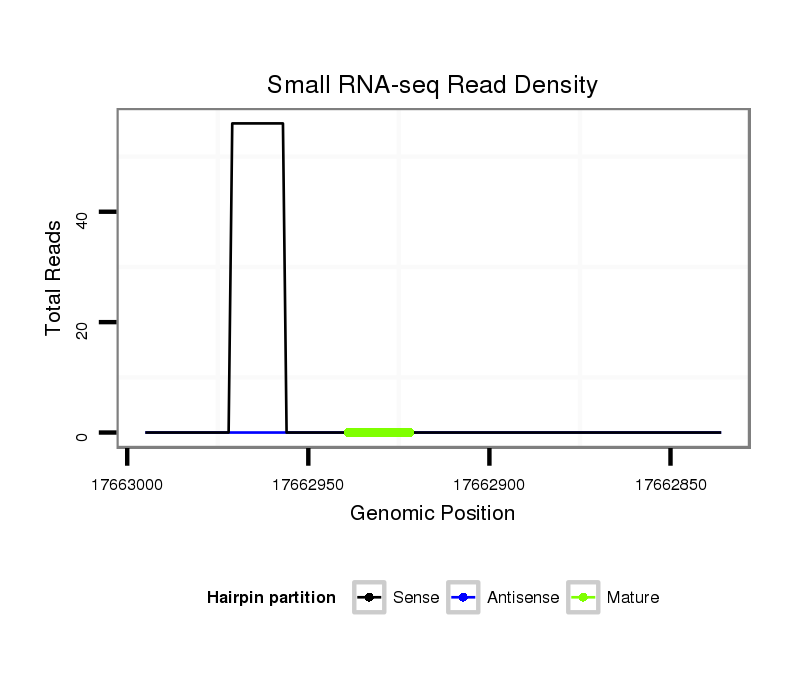
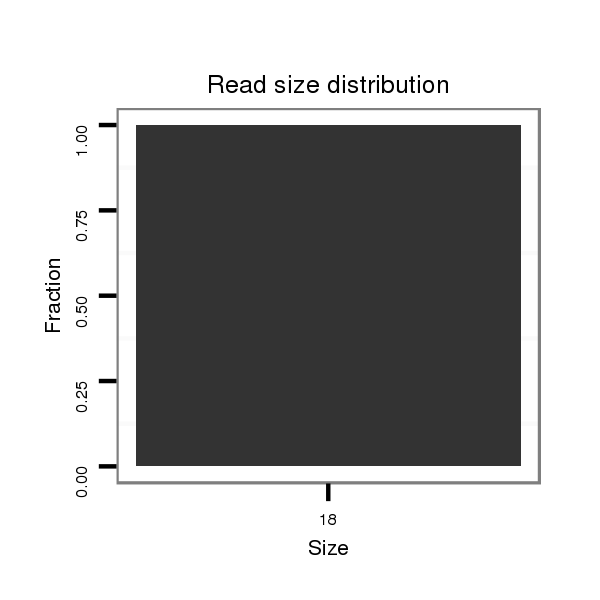
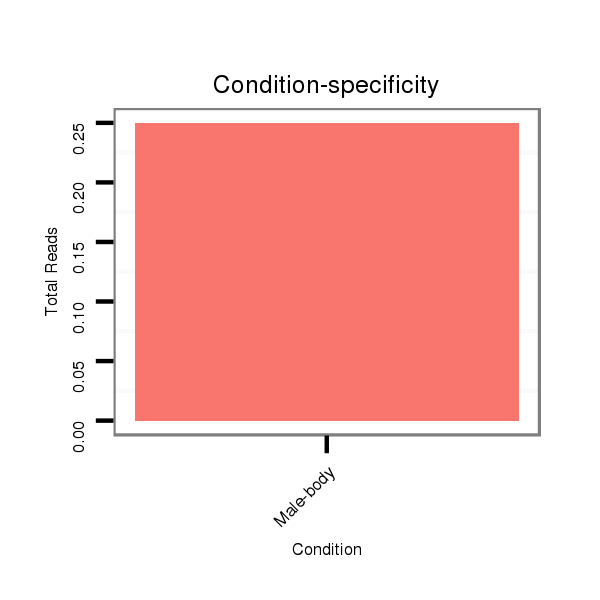
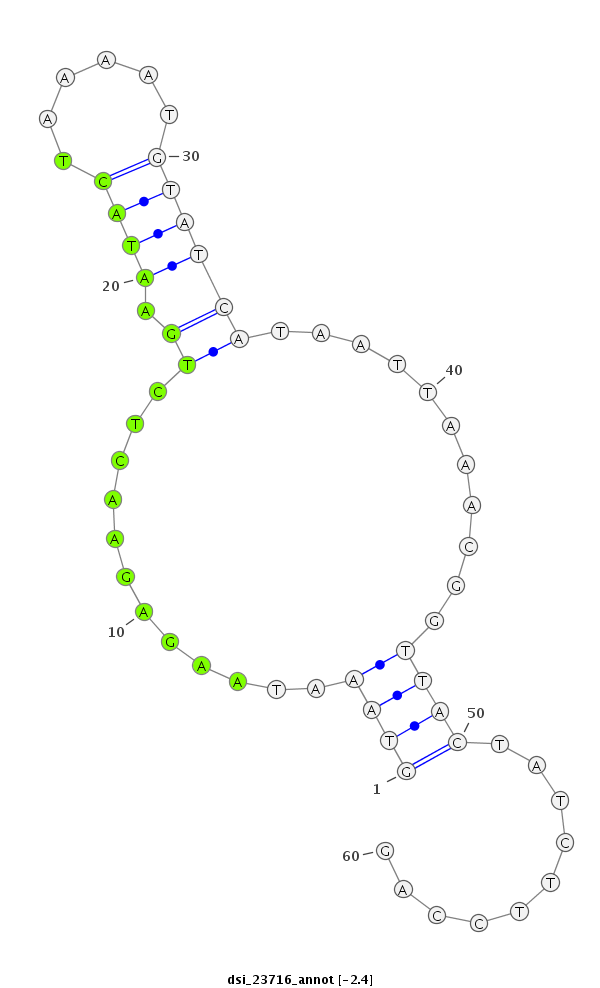
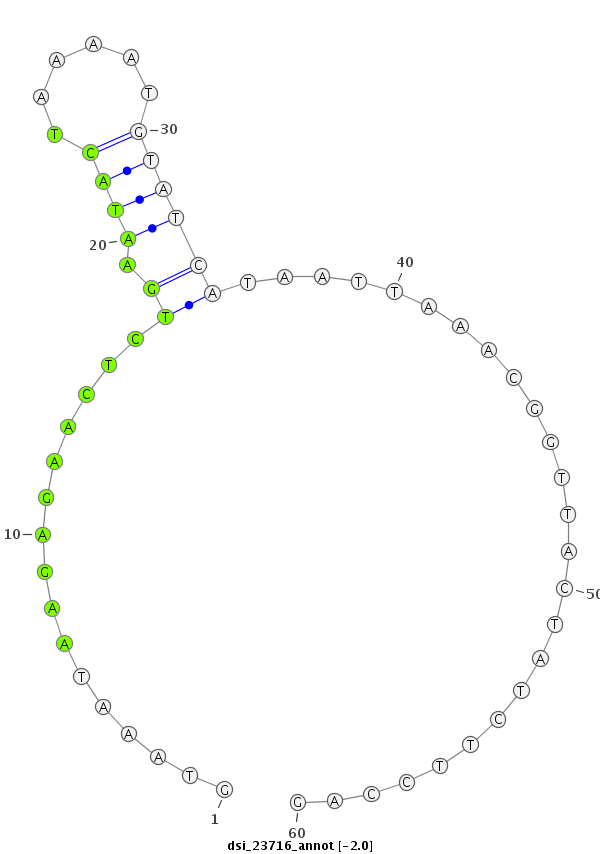
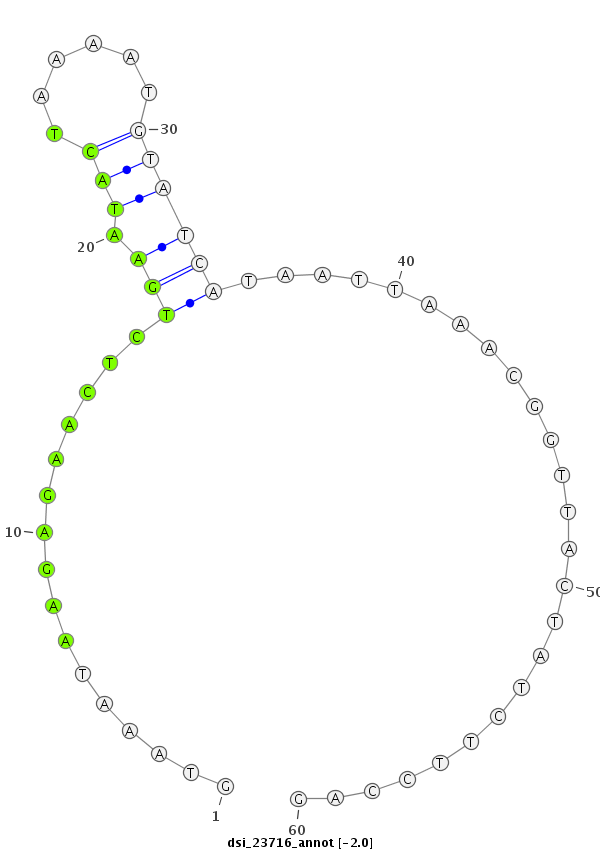
ID:dsi_23716 |
Coordinate:3r:17662886-17662945 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -2.4 | -2.0 | -2.0 |
 |
 |
 |
exon [3r_17662751_17662885_-]; CDS [3r_17662751_17662885_-]; CDS [3r_17662946_17663023_-]; exon [3r_17662946_17663023_-]; intron [3r_17662886_17662945_-]
No Repeatable elements found
| mature | star |
| ##################################################------------------------------------------------------------################################################## CGATGAAGCGACCGGAGCCGCATCAGGACTTTGAAAAGACCATACAGAGAGTAAATAAGAGAACTCTGAATACTAAAATGTATCATAATTAAACGGTTACTATCTTCCAGCTAAAGACACAGAAGGCGGCCCAGAAAAAGCCAGCCAGTGCTCCAATGGA **************************************************((((............(((.(((......))))))...........))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
M023 head |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................AGGACTTTGAAAAGA......................................................................................................................... | 15 | 0 | 1 | 56.00 | 56 | 46 | 8 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ........................................................ATGAGAACTTTGAATACTGAA................................................................................... | 21 | 3 | 5 | 6.80 | 34 | 0 | 0 | 20 | 13 | 0 | 0 | 1 | 0 | 0 |
| ........................AGGACTTTGAAAAGAG........................................................................................................................ | 16 | 1 | 7 | 4.29 | 30 | 21 | 5 | 0 | 0 | 2 | 0 | 0 | 2 | 0 |
| ..........................................................GAGAACTTTGAATACTGAA................................................................................... | 19 | 2 | 3 | 2.67 | 8 | 0 | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................AAGGACTTTGAAAAGA......................................................................................................................... | 16 | 1 | 15 | 2.20 | 33 | 21 | 9 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ........................................................ATGAGAACTTTGAATACTGA.................................................................................... | 20 | 3 | 13 | 1.15 | 15 | 0 | 0 | 11 | 3 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................GAGAACTTTGAATACTGA.................................................................................... | 18 | 2 | 3 | 1.00 | 3 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AAGAGAACTTTGAAGACTGAA................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................ATGAGAACTTTGAATACT...................................................................................... | 18 | 2 | 14 | 0.29 | 4 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................AAAAGAACTCTGAATA........................................................................................ | 16 | 1 | 12 | 0.25 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................AAGAGAACTCTGAATACT...................................................................................... | 18 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................AGAGAGGAAATAAGAAGACTC.............................................................................................. | 21 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................CTCTGAATACTAAACT................................................................................. | 16 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGAGAACTTTGAATACTGA.................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| .......................CAGGACTTTGAACTGACC....................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TCTGGACTTTGAAAAAA......................................................................................................................... | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| ........................................................ATGAGAACTCTGAAGACT...................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................TGAGAACTTTGAATACTGAA................................................................................... | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................TCAGGACTTAGAACTGACC....................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................TCAGGACTTTTAAAAAAC........................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................AGAGAGGAAATAAGAAGACT............................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GCTACTTCGCTGGCCTCGGCGTAGTCCTGAAACTTTTCTGGTATGTCTCTCATTTATTCTCTTGAGACTTATGATTTTACATAGTATTAATTTGCCAATGATAGAAGGTCGATTTCTGTGTCTTCCGCCGGGTCTTTTTCGGTCGGTCACGAGGTTACCT
**************************************************((((............(((.(((......))))))...........))))..........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|
| ...............TCGGCGGAGTCCCTAAACTT............................................................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................GGAGACCTATGATTCTACA............................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ..............CTCGGCGGAGTCCCTAAAC............................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................................................TTTTTCGGTCGGTCAAGT........ | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ...................TTTAGTCCGGAAACTTTTC.......................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 01:44 AM