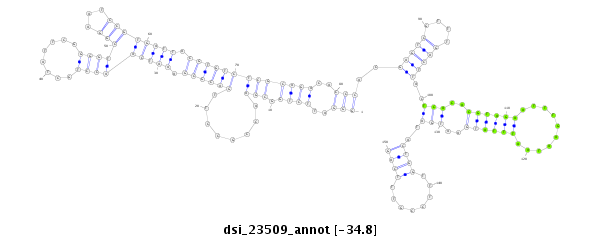
| ......................................................................................................................................................................................................TCGATTTTTTGATTA..................................... |
15 |
0 |
4 |
208.75 |
835 |
835 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................GTCGATTTTTTGATTA..................................... |
16 |
0 |
1 |
27.00 |
27 |
27 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................GTCGATTTTTTGATT...................................... |
15 |
0 |
1 |
13.00 |
13 |
13 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................GTCGATTTTTTGATTAG.................................... |
17 |
1 |
6 |
7.83 |
47 |
47 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................CAACCTTTAGGTCATGTGC.................... |
19 |
3 |
18 |
6.39 |
115 |
0 |
65 |
44 |
0 |
0 |
0 |
4 |
0 |
0 |
2 |
| .................................................................................................................................................................................................CCAAGTCGATTTTTTGATTA..................................... |
20 |
1 |
1 |
6.00 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................CAACCTTTAGGTCATGTGCT................... |
20 |
3 |
6 |
4.67 |
28 |
0 |
17 |
7 |
0 |
1 |
0 |
1 |
2 |
0 |
0 |
| ....................................................................................................................................................................................................AATCGATTTTTTGATTA..................................... |
17 |
1 |
5 |
2.40 |
12 |
11 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................CCAAGTCGATTTTTTGATT...................................... |
19 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................AACCTTTAGGTCATGTGC.................... |
18 |
2 |
2 |
1.50 |
3 |
0 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AAATCGATTTTTTGATTA..................................... |
18 |
1 |
2 |
1.50 |
3 |
1 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................ACCTTTAGGTCATGTGCT................... |
18 |
2 |
4 |
1.25 |
5 |
0 |
2 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................AATCGATTTTTTGATT...................................... |
16 |
1 |
16 |
1.13 |
18 |
18 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................AGTCGATTTTTTGATTAG.................................... |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AAGTCGATTTTTTGATTAG.................................... |
19 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................AACCTTTACGTCATGTGCTTTT................ |
22 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AGGTCGATTTTTTGATTA..................................... |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................TAAGTCGATTTTTTGATT...................................... |
18 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................CAAAGTCGATTTTTTGAT....................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................AACCTTTAGGTCATGTGCTT.................. |
20 |
3 |
4 |
1.00 |
4 |
0 |
2 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................ATCGATTTTTTGATTAA.................................... |
17 |
1 |
6 |
0.83 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................GGTCGATTTTTTGATTA..................................... |
17 |
1 |
4 |
0.75 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................TGTCGATTTTTTGATTA..................................... |
17 |
1 |
8 |
0.75 |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................GCCCCGTCGATTTTTTGATTA..................................... |
21 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AAAATCGATTTTTTGATTA..................................... |
19 |
1 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTAA.................................... |
16 |
0 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTAAGGTCTA.............................. |
22 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................ACCTTTACGTCATGTGCTT.................. |
19 |
2 |
2 |
0.50 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................ACCTTTAGGTCATGTGCTT.................. |
19 |
3 |
20 |
0.35 |
7 |
0 |
2 |
3 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTAGC................................... |
17 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................ATGTCGATTTTTTGATTA..................................... |
18 |
1 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AAGTCGATTTTTTGA........................................ |
15 |
0 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................TTATGACCAAAGTCAAAGTTGAT............................................... |
23 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................CCAAGATCGATTTTTTGATTA..................................... |
21 |
3 |
7 |
0.29 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................AAAATCGATTTTTTGATTAG.................................... |
20 |
2 |
4 |
0.25 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................AATCGATTTTTTGAT....................................... |
15 |
1 |
20 |
0.25 |
5 |
5 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................CAGTCGATTTTTTGATTAG.................................... |
19 |
2 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................GTCGATTTTTTGATCA..................................... |
16 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATCA..................................... |
15 |
1 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................CAACCTTTACGTCATGTG..................... |
18 |
2 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................................CCTTTAGGTCATGTGCT................... |
17 |
2 |
13 |
0.15 |
2 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATGA..................................... |
15 |
1 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................GCGATTTTTTGATTA..................................... |
15 |
1 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................ATGTCGATTTTTTGATT...................................... |
17 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................CGTCGATTTTTTGATTA..................................... |
17 |
1 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AAATCGATTTTTTGAT....................................... |
16 |
1 |
16 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTACCGT................................. |
19 |
2 |
8 |
0.13 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ................................................................................................................................................................................................CCCAAGTCGATTTTTTGA........................................ |
18 |
2 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................GGTCGATTTTTTGATT...................................... |
16 |
1 |
16 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................ACCTTTAGGTCATGTGC.................... |
17 |
2 |
16 |
0.13 |
2 |
0 |
0 |
0 |
0 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................CCAGTCGATTTTTTGAT....................................... |
17 |
2 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................TGTCGATTTTTTGATT...................................... |
16 |
1 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................TGTCGATTTTTTGAT....................................... |
15 |
1 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................AGATCGATTTTTTGATTA..................................... |
18 |
2 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................GCCGATTTTTTGATTA..................................... |
16 |
1 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................CAAATCGATTTTTTGATTA..................................... |
19 |
2 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................ATCGATTTTTTGATTAGCCG................................. |
20 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................CCCTGTCGATTTTTTGATTA..................................... |
20 |
3 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTGA.................................... |
16 |
1 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..............................................................................................................................................ACTGAATGAGAGTGTAT........................................................................................... |
17 |
2 |
16 |
0.06 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
| ................................................................................................................................................................................................ACCAAGTCGATTTTTTG......................................... |
17 |
2 |
16 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTGAGCAT................................ |
20 |
3 |
18 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................ACAATCGATTTTTTGATT...................................... |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTCTGATTA..................................... |
15 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TCGATTTTTTGATTG..................................... |
15 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................ACTTCGATTTTTTGATTA..................................... |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................GGAGTCGATTTTTTGAT....................................... |
17 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................CAACCTTTATGTCATGTGC.................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .................................................................................................................................................................................................CTGAATCGATTTTTTGATTA..................................... |
20 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................ATCGATTTTTTGATTAAA................................... |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ................................................................................................................................................................................................CCAAGATCGATTTTTTGAT....................................... |
19 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................................TAGATTTTTTGATTA..................................... |
15 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |