
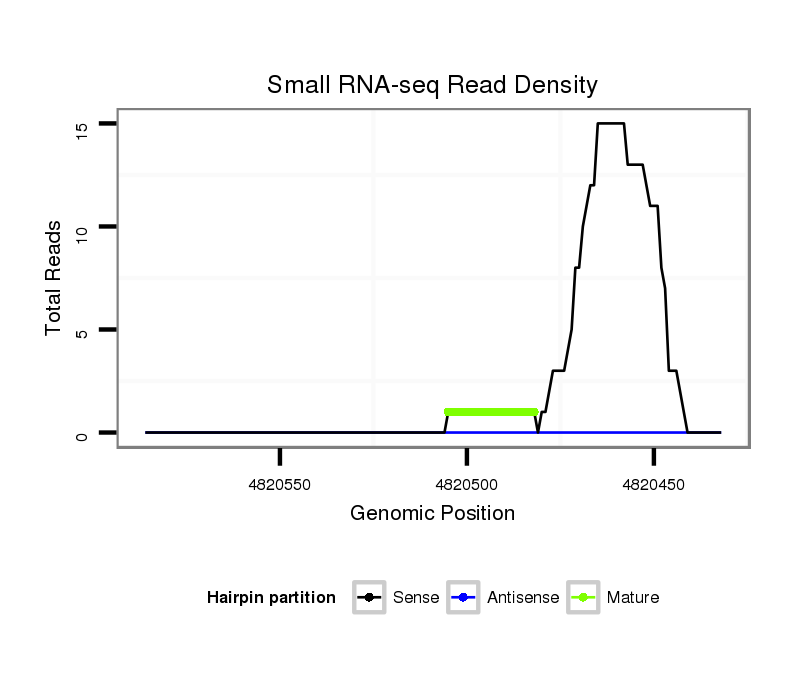

ID:dsi_23267 |
Coordinate:2r:4820482-4820536 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -1.3 | -1.3 | -1.3 |
 |
 |
 |
exon [2r_4820405_4820481_-]; CDS [2r_4820405_4820481_-]; CDS [2r_4820537_4821374_-]; exon [2r_4820537_4821374_-]; intron [2r_4820482_4820536_-]
No Repeatable elements found
| ##################################################-------------------------------------------------------################################################## CAAATCGGCGTTAACCAGGCCAAAGATTCCGCCACCAGGACCGCCATCAGGTGAGTGTCTTACCAGCCAACTAAATTCCAGTTTTTTTAATATTTTGATCGACAGAGCGCGAAATATCAAACGGACAGTCGAACGAGAGCATCAGTTCCATGGAC **************************************************(((((...)))))..........................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M025 embryo |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
M053 female body |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................ATCAAACGGACAGTCGAACGAGAGC............... | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGGACAGTCGAACGAGAG................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CAAACGGACAGTCGAACGAGAGCATC............ | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGGACAGTCGAACGAGAGCATCA........... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GCGCGAAATATCAAACGGACAGT.......................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................GCGAAATATCAAACGGACAGT.......................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ATCAAACGGACAGTCGAACGAGA................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................CAAACGGACAGTCGAACGAGAGC............... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................ATATCAAACGGACAGTCGAACGAGA................. | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TTTTTTTAATATTTTGATCGACAG.................................................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGGACAGTCGAACGAGAGCATCAG.......... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TATCAAACGGACAGTCGAAC..................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................ATCAAACGGACATTCGAACGAGAGC............... | 25 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................AAACGGACAGTCGAACGAGAGC............... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CGAAATATCAAACGGACAGTCGAACG.................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................AACGGACAGTCGAACGAGA................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GCTATATTTTGATCGACAGAGC............................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CGGACAGTCGAACGA................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......GCGTTAACAATGCGAAAGATT............................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................TTCCAGTTTTTTTAAAA............................................................... | 17 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...ATCGGCGTTGACCGGGC....................................................................................................................................... | 17 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTTTAGCCGCAATTGGTCCGGTTTCTAAGGCGGTGGTCCTGGCGGTAGTCCACTCACAGAATGGTCGGTTGATTTAAGGTCAAAAAAATTATAAAACTAGCTGTCTCGCGCTTTATAGTTTGCCTGTCAGCTTGCTCTCGTAGTCAAGGTACCTG
**************************************************(((((...)))))..........................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................TAAGGCGGTGGACCTG.................................................................................................................. | 16 | 1 | 4 | 0.50 | 2 | 1 | 1 | 0 | 0 | 0 |
| ..........................AAGGCGGTGGACCTG.................................................................................................................. | 15 | 1 | 13 | 0.23 | 3 | 3 | 0 | 0 | 0 | 0 |
| .........................TAAAGCGATGGTCCTGG................................................................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................TCGACTCACACAATGGT.......................................................................................... | 17 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................CCACTCCGAGAATGGTCGA....................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................AAGGCGGTGGTCCCG.................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................GTAGGCCACTCATAGAAG............................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/23/2015 at 06:13 AM