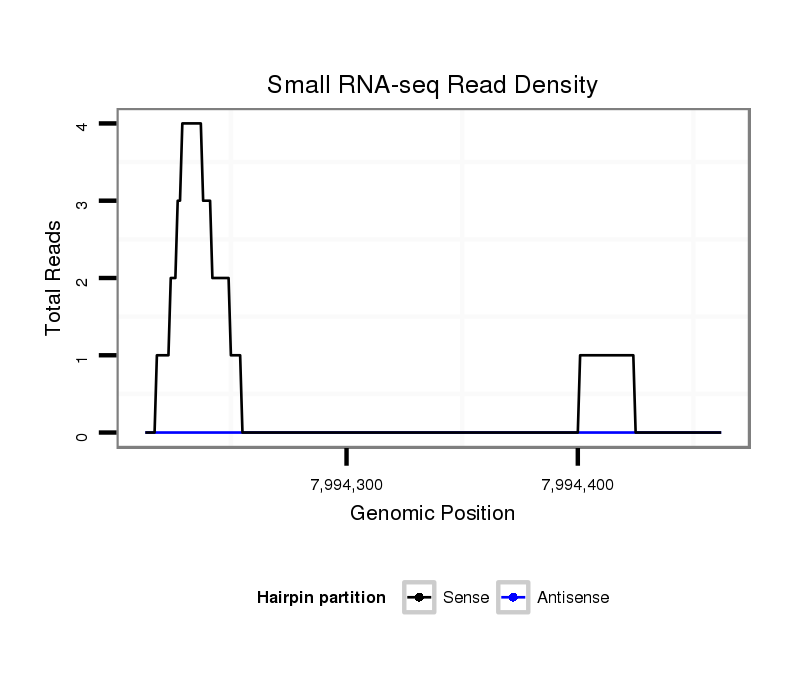
ID:dsi_23026 |
Coordinate:3l:7994263-7994412 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
CDS [3l_7994173_7994262_+]; exon [3l_7994069_7994262_+]; Antisense to three_prime_UTR [3l_7994306_7994355_-]; Antisense to CDS [3l_7994356_7995110_-]; Antisense to exon [3l_7994306_7995110_-]; intron [3l_7994263_7995970_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CGTGGCGCGAGCGCTGGACGGACTCCCAGTGGGGCATCAACATAAAAAAGGCAAGGCAACCCAAGCAAATTAACACACCGAACAATCAATATTCGTGTTTACCAATATATTGTAAATGCAATGAGGCCTGTACATTGAGAATTTTAGGGGTTCAAGTTGCCCGTCTCCGTGCTTTTGGTCTTGGGCTCGAAAACCGGATCCTCTGCCTGGGACAGGGCCAAAACCAGTTGGCGCTGCTTTACGGAGAGAT **************************************************....((((((..................................((((((((.........))))))))..((((.(((.................))).)))).))))))....((((...((((((((...))).)))))..))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M023 head |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................GAAAACCGGATCCTCTGCCTGGGA...................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............TGGACGGACTCCCAGTGGGGCATCAACA................................................................................................................................................................................................................ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...GGCGCGAGCGCTGGACGGATTT................................................................................................................................................................................................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........CGCTGGACGGACTCCCAG............................................................................................................................................................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................GACGGACTCCCAGTGGGGCAT..................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....CGCGAGCGCTGGACGGACTC................................................................................................................................................................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CGAATCCCGCATCCTCTGCC........................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................TTAGGGGTTCGAGTC............................................................................................ | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................CAATAGATTGTAAAT..................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................................................TTTTGGTCTTGGGAT............................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................TTCAAGTGACCCGTCT.................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ---------------------------------------------------------------------------------------------############################################################################################################################################################# GCACCGCGCTCGCGACCTGCCTGAGGGTCACCCCGTAGTTGTATTTTTTCCGTTCCGTTGGGTTCGTTTAATTGTGTGGCTTGTTAGTTATAAGCACAAATGGTTATATAACATTTACGTTACTCCGGACATGTAACTCTTAAAATCCCCAAGTTCAACGGGCAGAGGCACGAAAACCAGAACCCGAGCTTTTGGCCTAGGAGACGGACCCTGTCCCGGTTTTGGTCAACCGCGACGAAATGCCTCTCTA **************************************************....((((((..................................((((((((.........))))))))..((((.(((.................))).)))).))))))....((((...((((((((...))).)))))..))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
M023 head |
M024 male body |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................CACTCACGTTACTCCAGACAT...................................................................................................................... | 21 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................CGGGTCATGTAACTCTTA............................................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................TGTTAGTTATGAGCACTA....................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................GGCAGAGTCACAAACACCAG...................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................TTTGGCCTAGCAGCCTGA.......................................... | 18 | 3 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AAGTTCAACGTGCAG..................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................CTCCGTTGGGTTCGT....................................................................................................................................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........GCGACCTGCGTGATGGT.............................................................................................................................................................................................................................. | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 10:36 AM