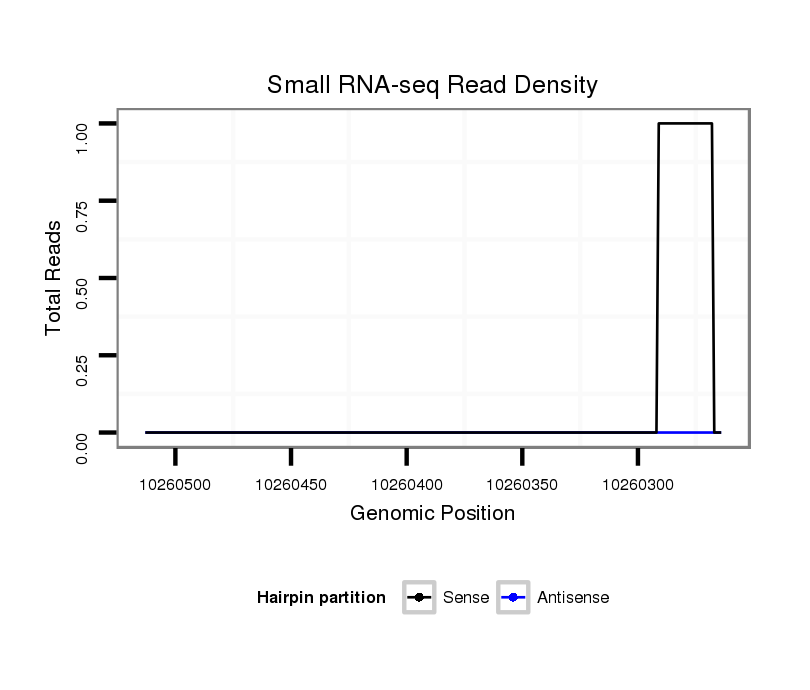
ID:dsi_22953 |
Coordinate:3r:10260314-10260463 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
ATGTTGTTCCTGGTACTCCCGGTTGCCCCAACACTCCTCAATATCCTACAGTGGTTCCGAATAATCCATCTAATCCCAATGTAAATGTTGTTCCTGGTACTCCCGGTTGTCCCAACACTCCTCAATATCCTACAGTGGTTCCGAATAATCCATCTAATCCCAATGTAAATGTTGTTCCTGGTACTCCCGGTTGCCCCAACACTCCTCAATATCCTACAGTGGTTCCAGATAAGCCACTGTGTCCCAATAC
**************************************************((((...........)))).......((((......)))).....((((...((((..((.(((........((((((..((((.(((....((........)).....))).)))).))))))....))).))..)))).)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................TTCCAGATAAGCCACTGTGTCCCA.... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................ACTCCCGGTTGCCGCACCA................................................. | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ..............ACTCCCGGTTGCCGCACCA......................................................................................................................................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................CAACGTTGATCCTGGTAC...................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................................GTAAACCGCTGTGTCCCA.... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................CAACGTTGATCCTGGTAC.................................................................. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
TACAACAAGGACCATGAGGGCCAACGGGGTTGTGAGGAGTTATAGGATGTCACCAAGGCTTATTAGGTAGATTAGGGTTACATTTACAACAAGGACCATGAGGGCCAACAGGGTTGTGAGGAGTTATAGGATGTCACCAAGGCTTATTAGGTAGATTAGGGTTACATTTACAACAAGGACCATGAGGGCCAACGGGGTTGTGAGGAGTTATAGGATGTCACCAAGGTCTATTCGGTGACACAGGGTTATG
**************************************************((((...........)))).......((((......)))).....((((...((((..((.(((........((((((..((((.(((....((........)).....))).)))).))))))....))).))..)))).)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M053 female body |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902008 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................ACAGTTACAACCAGGTCCA........................................................................................................................................................ | 19 | 3 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................ACAGTTACAACCAGGTCCA.................................................................... | 19 | 3 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 | 0 |
| ......................................GTTATAGCATGTCACCAACAC............................................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GTTATAGCATGTCACCAACAC........................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................TCAGGGATGTGAGGAGT.............................................................................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................TGTAACCAAGGCTCATCAG........................................................................................................................................................................................ | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TGTAACCAAGGCTCATCAG.................................................................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................TAGGTGTATTAGGGTTAC......................................................................................................................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................TAGGTGTATTAGGGTTAC..................................................................................... | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................GGCCAACAGGTTTGTAA................................................................................................................................... | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................ACCATGGGGGCCAGC......................................................... | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................ACCATGGGGGCCAGC............................................................................................................................................. | 15 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................ACCAAGGCTGATAAGGTCG.................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................ACCAAGGCTGATAAGGTCG................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/24/2015 at 10:39 AM