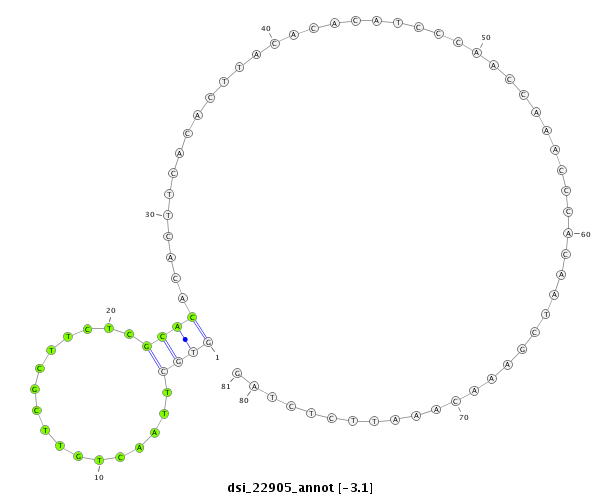
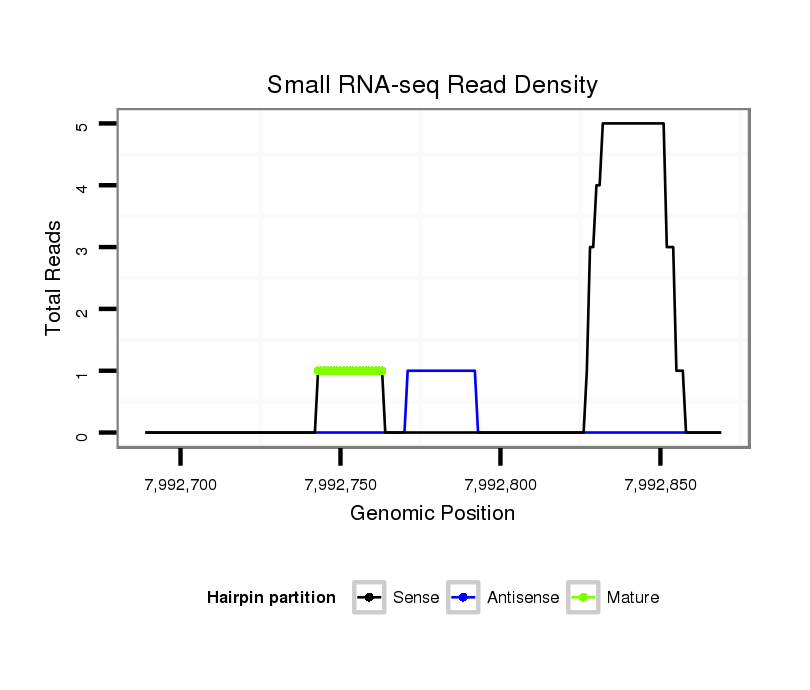
ID:dsi_22905 |
Coordinate:3l:7992739-7992819 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
CDS [3l_7992611_7992746_+]; three_prime_UTR [3l_7992747_7993497_+]; exon [3l_7992611_7993497_+]
No Repeatable elements found
| ##################################################################################################################################################################################### AAAGAGGCACGACCTGCGGCTGGAGGACCACTATGCTATGGATATCAAAGGTGCTTAACTGTTCGCTTCTCGCACACACTTCACACTTACACACATCCCAACCAAACCCACAATCGAAACAAATTCTCTAGTTGATTCACCTCAAACTCTAGGCCTGAACAGTGGCGTGAGCATTGGCGGA **************************************************((((.................))))........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
M025 embryo |
SRR902008 ovaries |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........ACCTGCGGCTGGATCACC........................................................................................................................................................ | 18 | 2 | 9 | 4.44 | 40 | 28 | 0 | 0 | 0 | 7 | 4 | 1 | 0 |
| ...........ACCTGCGGCTGGATCACCCC...................................................................................................................................................... | 20 | 3 | 5 | 1.80 | 9 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAACTCTAGGCCTGAACAGTGGC............... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CCTCAAACTCTAGGCCTGAACAGTGGC............... | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TCAACCTCTAGGCCTGAACAGTGGCGTG............ | 28 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................TCAAACTCTAGGCCTGAACAGTGGCGTG............ | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CCTCAAACTCTAGGCCTGAACAGT.................. | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ACCTCAAACTCTAGGCCTGAACAGT.................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TTAACTGTTCGCTTCTCGCAC.......................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................GCTATTGATATAAAAGGTG................................................................................................................................ | 19 | 2 | 3 | 0.67 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .............CTGCGGCTGGAGGAC......................................................................................................................................................... | 15 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............CCTGCGGCTGGATCACC........................................................................................................................................................ | 17 | 2 | 20 | 0.15 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........ACCTGCGGCTGGATCACCC....................................................................................................................................................... | 19 | 3 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................AATTCTCTGGTTGACTCA.......................................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
TTTCTCCGTGCTGGACGCCGACCTCCTGGTGATACGATACCTATAGTTTCCACGAATTGACAAGCGAAGAGCGTGTGTGAAGTGTGAATGTGTGTAGGGTTGGTTTGGGTGTTAGCTTTGTTTAAGAGATCAACTAAGTGGAGTTTGAGATCCGGACTTGTCACCGCACTCGTAACCGCCT
**************************************************((((.................))))........................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
O001 Testis |
M053 female body |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................GAGGTGTGAAGGTATGTAGGGT................................................................................. | 22 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................ATGGTGATACGATACC............................................................................................................................................ | 16 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................ATGGTGATACGATACCT........................................................................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................GTAGGGTTGGTCTGGGT....................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................TGTGAATGTGTGTAGGGTTGGT............................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................GAGGTGTGAATATATGTAGGGT................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................GTAGGGTTGGTCTGGGTTT..................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................GAGCGTGTGTACAGTCTGA.............................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| ........................................................................................................................................................CGGACTTGTCCCCGGACTTG......... | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................TAGGGTTGTTTTGGTGGTTA................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................CTGTAGGGTTAGTTCGGG........................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................AAGGGATCAACCAATTGGA....................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................CCTGGTGATATGATAGCT........................................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 10:35 AM