| droSim2 |
3l:7123131-7123514 - |
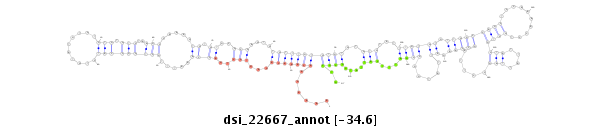
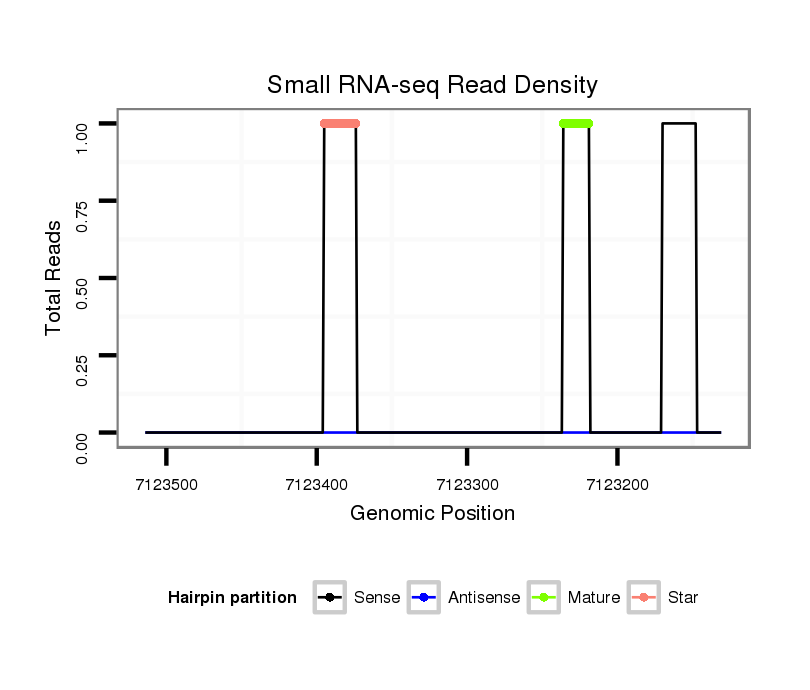
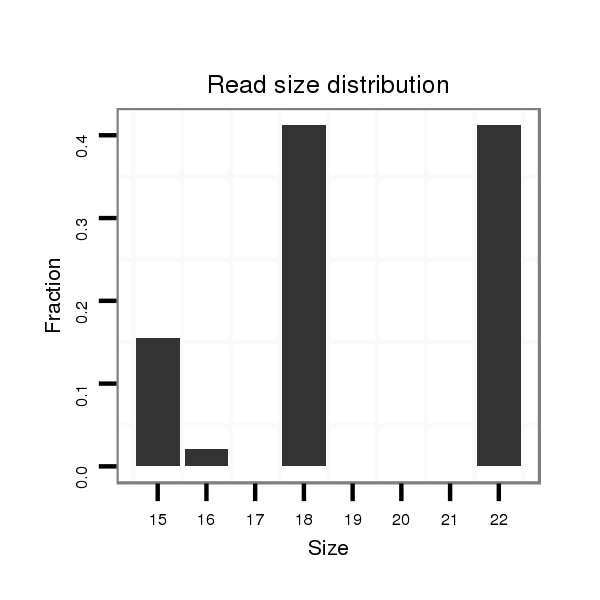
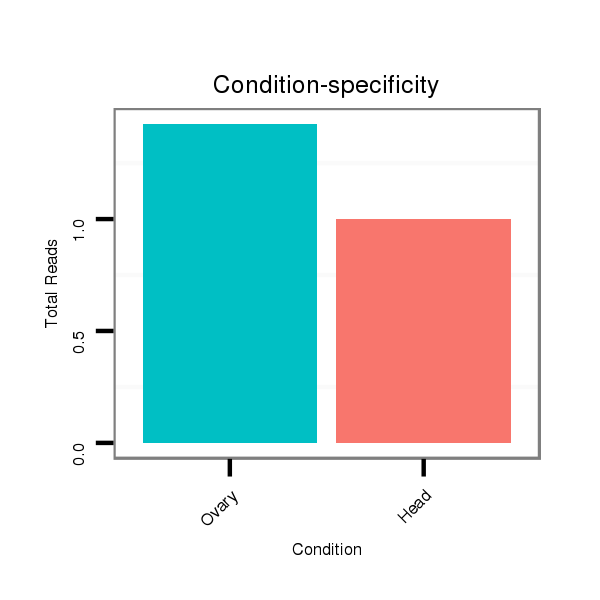
dsi_22667 |
T----------AT-CTGAGAT--AAATAACGCGCA-----------------------GCAAAAGCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAAAA----A--------AA--A-A---GAAAAAG-AA--AAGAACAA---ACAGAGTGCTG---------------------------------------------ACCAAATTTGCTGG-CCAT--CT-----------GT------A----T--GTGTGT---------------------------GTGTCTAAGCCAAAGT-----------------G--------TGTGCGCGAGTGTG--------TTT--------GTGTGTGTGTGTATTTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGCCGC-------------------------------------------------AAAAAGCGAAAC-----------AAAAATGTAAAA---C---AGCAGT--GCCCGGGCCA-----AGAAGATT--------T----GAGCCAAG-TTAGACG-AGTT------------------------ |
| droSec2 |
scaffold_2:7182568-7182947 - |
|
T-------------CTGAGAT--AAATAACGCGCA-----------------------GCAAAAGCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAAAA----A--------AA--A-A---GAAAAGG-AA--AAGAACAA---ACAGAGTGCTG---------------------------------------------ACCAAATTTGCTCG-CCAT--CT-----------GT------A--------TATGT---------------------------GTGTCTAAGCCAAAGT-----------------G--------TGTGCGCGAGTGTG--------TTT--------GTGTGTGTGTGTATTTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGACGC-------------------------------------------------AAAAAGCGAAAC-----------AAAAATGTAAAA---C---AGCAGT--GTCCGGGCCA-----AGAAGATT--------T----GAGCCAAG-TTAGACG-AGTT------------------------ |
| dm3 |
chr3L:7256517-7256892 - |
|
A----------AT-CTGAGAT--AAATAACGCGCA-----------------------GCAAAAGCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGACGTGAAGTATACGCCCGATCTGTTGGCGAGAA----A--------AA-TA-A---GAAAAAG-AA--AAGAACAA---ACAGAGTGCTG---------------------------------------------GCCAAATTTGCTCG-CCAT--CT-----------GT------A----TGTGTCT-----------------------------GTGTCTAAGCCAAAGT-----------------G--------TGT--GCGAGTGT--------------------GTTTGTGTGTGTATTTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGCCGC-------------------------------------------------AAAAAGCGAAAC-----------AAAAATGTAAAA---C------AGT--GCCTGGGCCA-----AGAAGATT--------T----GAGCCGAG-TTAGACG-AGTT------------------------ |
| droEre2 |
scaffold_4784:4086436-4086817 + |
|
A----------AT-CTGAGAA--AAATAACGCGCA-----------------------GCAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTT--------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAGAA----ATAGCCGAAGAAT----AA-------A--AA-AG----A---ACAGAGTGCTG---------------------------------------------GCCAAATTTGCTCA-CCATTTCT-----------GT------A--------TGTGTGTGTGTGTGAGTGTGTGGTGTGCGTGTGTGTCTAAGCCAAAGT-----------------------------------------------------------------GTGTGTATTTGT--------------------------------------GTGTTG--------------------TCCAAC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGATGCAATTTGCTGTGCCGCCGC-------------------------------------------------AAAAAGCGAAAC-----------AAAAATGTAAAA---C------AGA--GCTTAGGCCG-----AGAAGATT--------T----GAGCTGAG-TTAGACG-AGTT------------------------ |
| droYak3 |
3L:7858407-7858804 - |
|
GCAAACGAAAAAT-CTGAGAA--AAATAACGCGCA-----------------------ACGAAATCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAGAA----TTAACCAACGAAT----AA-------A--AA-AGAACAA---ACAGAGTGTTG--------------------------------------------GGCCAAATTTGCTCA-CCAT--CTA----CG----GT----GT--GTGT--GTGTGT---------------------------GTGTCTAAGCCAAAGT-----------------G--------TGT--GCGAGTGTGT------------------GTTTGTGTGTATATTTGT--------------------------------------GTGTTG--------------------TCCAAC----------------------TTCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGATGCAATTTGCTCTGCCGCCGC-------------------------------------------------AAAAAGCGAAAC-----------AAAAATGTAAAA---C------AGA--GCTTAGGCCG-----AGAAGATT--------T----GAGCTGAG-TTAGACG-AGTT------------------------ |
| droEug1 |
scf7180000409006:215867-216213 + |
|
A----------A----------------------------------------------------TCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTAAACGCCTAACCCGCTGGCGAGAA----A--------AAAT----AA-------A--AC-AGAACAA---ACAGAGTGCTG---------------------------------------------GCCAAATTTGCTCA-CCAT--CT-----------GT------A----T--ATCT-----------------------------GTGTCTAAGCCAGAGT-----------------T--------TGT-CGT---TGTG--------TCTATGTGTGTGTGTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGCCGC-------------------------------------------------AAAAA---TACC-----------AAAAA---AAAACAATATAAGCAAA--GTTTAGGCCG-----AGAAGATT--------T----GAGCTAAG-TTAGACG-AGTT------------------------ |
| droBia1 |
scf7180000302193:6175487-6175819 - |
|
G---------------------------GCGCGCA-----------------------GTAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTTTACGCCCGATCTGCTGGCCAGCG--------------GGA----AA-------A--AC-AG----A---ACAGAGTGCTG---------------------------------------------GCCAAAATTGCTCA-CCAT--CT-----------GT------A--------TCT-----------------------------GTGTCTAAGGCAG--T-----------------G------------------TG--T--------------GCTTGTGTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGAGCCGCCGC-------------------------------------------------ACAA------AC-----------AAAAATA--AAAAAACTGAAACCGA--GCTTAGGCCG-----AGAAGATT--------T----GAGCTTAG-TTAGACG-AGTT------------------------ |
| droTak1 |
scf7180000415773:421609-421944 + |
|
C----------A-----------AAAAAACGCGCA-----------------------GCAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGGCAGAA----A--------AA--AATGGA-------A--AC-AG----A---ACGGAGTGCTG---------------------------------------------GCCAAATTTGCTCA-CCAT--CT-----------GT------A--------TCT-----------------------------GTGTCTAAGCTAA-GT-----------------G--------------------------------------TGTGTTTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGCCGC-------------------------------------------------AAAAA---AACC-----------AAAAAAGAAAAA---C------CGA--GCTTAGGCCG-----AGAAGATT--------T----GAGCTTAG-TTAGACG-AGTT------------------------ |
| droEle1 |
scf7180000491255:165938-166296 + |
|
C----------AA----------ACAAAACGCGTA-----------------------GCAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAGAA----A--------AA--A------------A--AT-AG----A---ACAGAGTTCCG---------------------------------------------TCCAAATTTGCTCA-CCAT--CT-----------GT------A--------TCT-----------------------------GTGTCTAAGCCAGAGTTTT--CTATATGTA----------------TGTGTGTG--TGTTCGAGTGA----GTGTGTTGG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGAGCCGCCGC-------------------------------------------------AAAAA-----AC-----------AAAA-----AAA---C------CAATGGTTTGGGCCG-----AGAAGATT--------T----GAGCTTAG-TTAGACG-AGTT------------------------ |
| droRho1 |
scf7180000766916:63050-63422 - |
|
A----------C----GAAAACACAAAAACGGGCA-----------------------GCAAATTCAAC-ATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCTGCTGGCGAGAA----A--------GA--AA---A-------A--AT-AG----A---ACAGAGTGCTG---------------------------------------------GCCAAAATTGCTCA-CCAT--CT-----------GT------A--------TCT-----------------------------GTGTCTAAGCCAGAGCTTTGCCTGTGTGT------------------GCGAGTGTGTGTGCC----------AGTGTGTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGTCCATTTCATTATGGCGCTGCAATTTGCTGAGCCGCCGC-------------------------------------------------AGAAA---A-ACACAAAAACGCCAAA----AAAAA---T------AGA--GTTTGGGCCG-----AGAAGATT--------T----GAGCTTAG-TTAGACG-AGTT------------------------ |
| droFic1 |
scf7180000453839:1262667-1263014 - |
|
A----------C-----------AAAAAACGCGCA-----------------------GTAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTAAACGCCCGACTTG--------AA----A--------AA--AA---A-------AAC--AAGAACAA---ACAGAGTGCTG---------------------------------------------GCCAAATTTGCTCA-CCAT--CT-----------GT------A--------TCT-----------------------------GTGTCTAAGCCAGAGT-----------------T--------TTGTCGTGTGTG--T--------------GTGTGTGTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTTCCGCCGC-------------------------------------------------AAAAAAAAAAAT-----------ACAAAAAAAAAA---C------TAA--CTTTAGGCCG-----AGAAGATT--------T----GAGCTAAG-TTAGACG-AGTT------------------------ |
| droKik1 |
scf7180000302329:36377-36705 - |
|
A-----------------------AAAAGCGCGCA-----------------------GTAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTGAAGTATACGCCCGATCGGCTGGCGAGAA----A--------AA--A---------------AC-AGAACAAACAACGGAGTGTTG---------------------------------------------GCCAAATTTGCTAA-CCAT--CT-----------GT------AAATGT--ATCT-----------------------------GTGTCTAAGCCAGAGT-----------------C--------TGTATCCGAG----------------------TGTGTG--------TGTGT--------------------------------------GTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCCGCCGC---------------------------------------------------------------------------------------C------AGA--GTTTGGGCCG-----AGAAGATT--------T----GAGCTTAG-TTAGACG-AGTT------------------------ |
| droAna3 |
scaffold_13337:14712886-14713222 - |
|
A----------AC-C--------AACAATCGCGCA-----------------------GTAAATTCAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCATGAAGTATACGCCCGATCTGTAAGCGAGAA----A--------AA--A---------------AA-AGAACAA---ACAGAGTATTG---------------------------------------------GCCAAATTTGTTAG-CCAT--CT-----------GT----GC--TAGT--ATCTGT---------------------------GTGTCTAAGCCAGAGT-----------------G--------TACTTAAGAGTGTG--------TGT--------GCTTGTGTGTGTGTGTGT--------------------------------------GTGTGG--------------------G---------------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGGGCAGCC-------------------------------------------------------------------------------------------------------------CGAAAACAGAAGATT--------TTTGCGAGCTTAGCTTAGACG-GGTT------------------------ |
| droBip1 |
scf7180000396390:208569-208892 - |
|
T----------T----AAAAAAAAACAAGCGCGCA-----------------------GTAAATTAAACTATTTTTAGCGGATTACGTGAGTGTCATTTGAATTTT-------------TTCGGCGTCAAGTATACGCCCGATCTGTAGGCAAGAA----A--------AA--A------------------AGAAAAA---TATGAGTATTG---------------------------------------------GGCAAATTTGTTAG-CCAT--CT-----------GT----GA--GAGT--TTCTGT---------------------------TTGTCTAAGCCAGAGT-----------------G--------TAT--CTAAGAG--------------------TGTTTG----------TGT--------------------------------------GTGTG-------------------------GG----------------------TCCAAAACCT--GCTGCTATTATTAGTCTTAGCCAAGATTATGGGCGATTTCATTATAGCGCTGCAATTTGCTGGGCAGCCCG-------------------------------------------------AAAA-------------------------------------------------------C-----AGAAGATT--------TTTGCCAGCATTGCTTAGACG-GGTT------------------------ |
| dp5 |
XR_group8:3347356-3347729 - |
|
-----------------------------CGCGCA-----------------------ATAAATTCCACTATTTTTAGCATATTACGTGAGTGTCATTTGAATTTG-------------TT--GCGTGAAGTGTAGCCCCGGTCCGGGGGCAA--------------------------------------AAGAACAA---ACAGAGTTATA---------------------------------------------CCCAAATTTGTTCA-CCAT--TTGGCTA-ACTCTGTGCCTGC--TTGT--CTCT------------------------------------------------------GTGTATGTG----------------------T------------------GTCTCTGTGTGCGAGTGT--------------------------------------CTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCTGCCGCCAGCCCCAGTCCCAGCCCCAGCTCCAGCCCCAGCTCCAGTCCCAGTCCCAGAA-----------------------------------------AGA--GTTTGGGCCG-----AGAAGATTTTAGAGTTT----GAGTTTGG-TTAGACG-AGTT------------------------ |
| droPer2 |
scaffold_65:151309-151694 - |
|
-----------------------------CGCGCA-----------------------ATAAATTCCACTATTTTTAGCATATTACGTGAGTGTCATTTGAATTTG-------------TT--GCGTGAAGTGTAGCCCCGGTCCGGGGGCAA--------------------------------------AAGAACAA---ACAGAGTTATA---------------------------------------------CCCAAATTTGTTCA-CCAT--TTGGCTA-ACTCTGTGGCTGC--TTGT--CTCT------------------------------------------------------GTGTGTGTGTGTCTCTGTGT--GTG--TG----------TGT--------GTGTGTGTGTGCGAGTGT--------------------------------------CTGTTG--------------------GCCAGC----------------------TCCAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGGCGATTTCATTATGGCGCTGCAATTTGCTGTGCTGCCGCCAGTCCCAGTCCCAGTCCCAGCCCCAGCTCCAGCTCC------AGTCCCAGAA-----------------------------------------AGA--GTTTGGGCCG-----AGAAGATTTTAGAGTTT----GAGTTTGG-TTAGACG-AGTT------------------------ |
| droWil2 |
scf2_1100000004511:9095055-9095322 - |
|
A----------AA-C---------GAAAAA----------------------------------------------------------------------------------------------------------------------GGAGAGAA----A--------AA--A---------------AT-------------------------------------------------------TGTTTCTAACAAGCAAAAATTGCTAAGCCCT--TTGGCTATAATGTGCATGTGT--TTGA--CTCT------------------------------------------------------GTGTGTGTG----------TGTGCGAA----------------------TGTGTG--------TGTGTGTGTTGGCGTCTTGATGTATTGGTGTGCTGTTGTATATGTGTTG--------------------GC---------------------------CGAAAACCT--GCTGCTATTATTAGCCTAAGCCAAGA-------------GATTATAAGGCTGCAATTTGCTGAAA----AC-------------------------------------------------AACAACAACAAC-----------AAAAATA----A---C------AAC--GAAAAAGCAG-----ACAAGTTT--------T----GCCC----------------------------------------- |
| droVir3 |
scaffold_13049:2394915-2395246 - |
|
A----------AACTTGAAAA---AAAAAAACACA----------TTTATATATACATATATATTCAACTATTTTTAGCGTATTACGTGAGTGTCATACGAATTTGGGCAGTTGAGCTTTTGAGC---------------GAG----TGTCGAGCGAAAAA--------A----------------------AAAACAA---AAAATGTGTTAAATAGCGTACACTGTAC---------AGCAAA-TGCTTCTAACAAGC-------GCT-G-CCCT--TT-----------GT----GG--GCG----TCT-----------------------------GT-----------------------GTGTGTGTG----------T--GTG--TG----------TGT--------GTGTGTGTGTGTGTATGT--------------------------------------CTGTAG--GCGCATAACTTAGCTTGAGCCAAG----------------------CCAAAAACCT--GCTGCTATTATTAGCCTTAGCCAAGATTATGGCCGATTTG------------------GC----------------------------------------------------------------------------------------------------------------------TG-----AAAAAA--------------------------AAATG-AGTT------------------------ |
| droMoj3 |
scaffold_6654:1484582-1484903 + |
|
A----------AC-TCGAAAA--AAAAAGTGAGCAGGCAGCAGCA-------ATAC--GTATATTCAACTATTTTTAGCGCATTACGTGAGTGTCATACGAATTTG-------------GCAGC--------------------TGAGCGCAAGCA----A--------CA--A---------------AA-AAAGCAAACACAAATGTGCTAAATAGTGTACACTGTACTACAGGCCCAACAAAGTCTTTCTAACAAGC-------GCT-G-CCCT--TT-----------GG------C--------TAT------------------------------------------------------GTGTGTGTG----------T------GTG--T--------------GTGTGTGTG--------TGTGT--------------------------------------GTGTTGAGGCGCATAACTCAGCTTGAGCCAAGCCAAAAAAAAAAAAAAACAAAATACAAAACCT--GCTGCTATTATTAGCTTAAGCCAAGATTATGGCCGATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_4789:4602-4781 - |
|
C----------GT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GT------GTG------TGT-----------------------------GTGTGTGTGCGTGTGT-----------------GTGTG----TGTGTGTGTGTG--T--------------GTTTGTGTG--------TGTGT----------------GTGTTTGT--------------GTGT-G--------------------CCAAGT----------------------TCTGGATTCTTTATCGCTCGTCTCATCCTCATCCAATG----GGGCAATGATGTTGCTTGGTTGCATGTCGCC----------------------------------------------------------------------------------------------------------------------------------------------------C-------AA--GAGGTCCTGGATAAAAAAGAGATGTAAAAC |