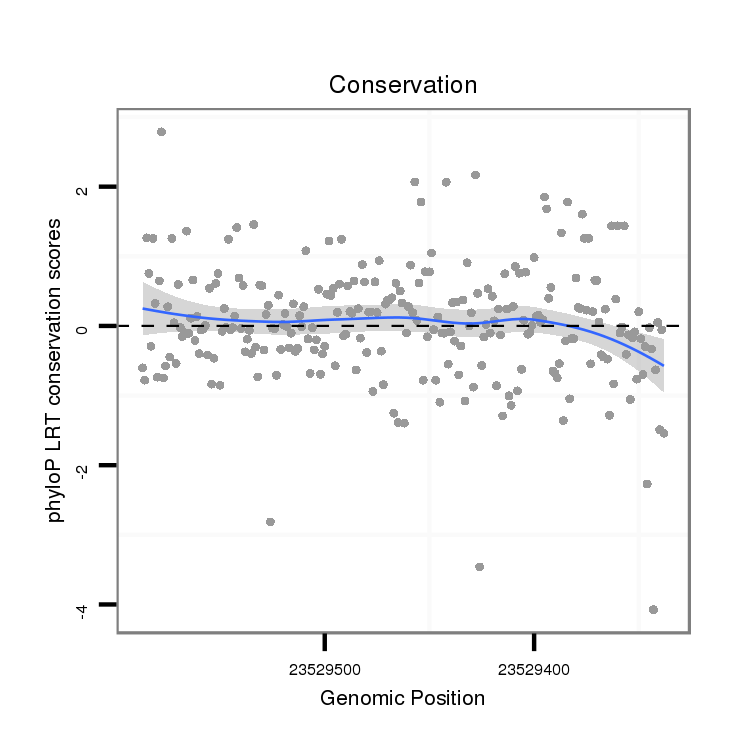
| AACAACAACAACAACAACTGTAATTGCAACTGCAACA------GC---AGCGG---CCACAAC---AACAACA---GCTCCAGATAGCTG----------CAGC------AGCAA---------------------------------------GAACATATTACAATATAT-A------------T-TTTATTTTTTTAATTTAATT-ATTTAA------AGTAAACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTATTT | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ...............................GCAACA------GC---AGCGG---CCACAAC---............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................ACAAC---AACAACA---GCTCCAGATAGC......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GG---CCACAAC---AACAACA---GCTCCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GTAAACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------A....................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AACAACAACAACAACAACTGTAAT......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTCCAGATAGCTG----------CAGC------AGCA............................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................TCCAGATAGCTG----------CAGC------AGCAA---------------------------------------GAAC................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................A---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATT.... | 28 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........CAACAACAACTGTAATTGCAACT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACAACAACAACAACTGTAATTGCAACC.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACAACAACAACAACTGTAATTGCAACT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................ACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GAT................................................................................................................................................................................. | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..CAACAACAACAACAACTGTAATT........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................CCAGATAGCTG----------CAGC------AGCAA---------------------------------------GAACAT................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................ACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------....... | 18 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................ATT-ATTTAA------AGTAAACAA---------------------AGAG--A-----.......................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...AACAACAACAACAACTGTAATTGCA..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................AG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GA.................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GTAAACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-.................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................AAC---AACAACA---GCTCCAGATAGCTG----------CAGC------................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AACAACA---GCTCCAGATAGCTG----------CAGC------A.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................CAACA------GC---AGCGG---CCACAAC---............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AACAACAACAACAACAACTGTA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................GCGG---CCACAAC---AACAACA---GCT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .ACAACAACAACAACAACTGTA........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................CAAC---AACAACA---GCTCCAGATAGC......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACAACAACAACAACTGTAATTGC...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................AATTGCAACTGCAACA------GC---AGCGG---CC....................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................GCTCCAGATAGCTG----------CAGC------AGCAA---------------------------------------GAACA.................................................................................................................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................AAACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GTAAACAA---------------------AGAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------.................................................................................................................................................................................... | 28 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................ACTGCAACA------GC---AGCGG---CCA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......ACAACAACAACTGTAATTGCAACT.................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................GAG--A-----AACAA-----------------------------------A-----------------------------------------AATGC-AAGC------------------------------------------GATA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------....... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....ACAACAACAACAACTGTAATTGCAAC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTCCAGATAGCTG----------CAGC------AGC................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTCCAGATAGCTG----------CAGC------AGCAA---------------------------------------G...................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................GTAATTGCAACTGCAACA------GC---................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GCAACTGCAACA------GC---AGCGG---C........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................ATTGCAACTGCAACA------GC---AGC.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AACAACAACAACAACAACTGT............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 21 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AACAACAACAACAACAACTG............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |