ID:dsi_22492 |
Coordinate:3r:4405524-4405673 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
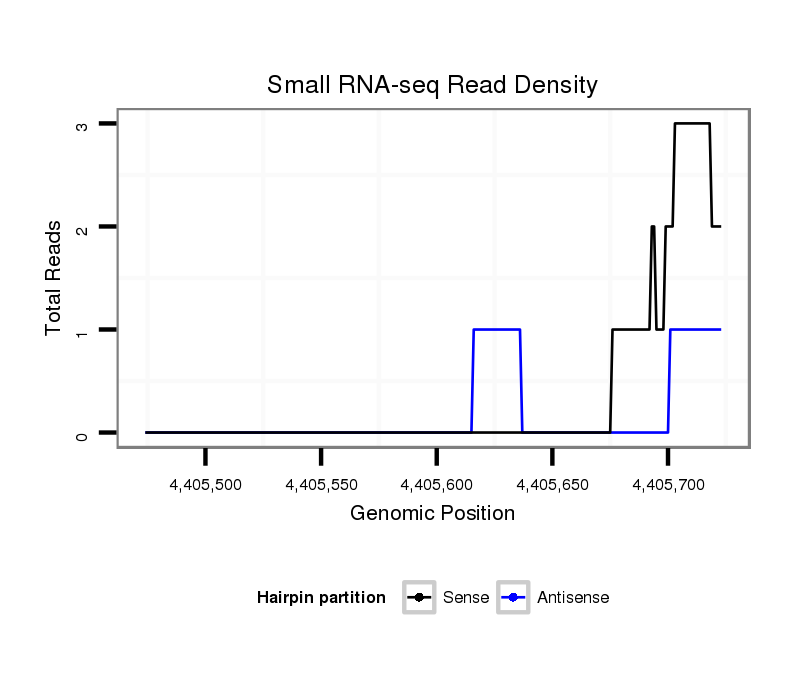
| Legend: | mature | star | mismatch in read |
 |
intergenic
No Repeatable elements found
|
TATGGAAAATGACACTGCCAAATTCGATTCCCAGATGAAAAGAGTTATGTACTTTGGGATCCCCAATGTGTGAAGCTATTGAAGAAAATTGGCATAGGTATAAAAAGCAGACATATAGCGATAAGAAGCGGCGATAAGATAGGTCTCCGTGCACGTTTTAAAACTGAAGTGTTAACTAATCACCATCACTTTCCCCGCAGCCAACGACTTGCAGCGCACTAAACTGACCTACGAGAGTCGCGCCAATCTG
**************************************************.....((((......(((.((((.((((...........))))..............(((......((((.......((((.(((.......))).))))...))))......))).(((....)))..))))))).....)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR902008 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TGAACTGTCAACTGATCACCATC............................................................... | 23 | 3 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................TGAACTGTCAACTGATCACCAT................................................................ | 22 | 3 | 1 | 4.00 | 4 | 2 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................GATAGGTCTCTGTGCAGGGTTTA.......................................................................................... | 23 | 3 | 3 | 1.67 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AACGACTTGCAGCGCACTA............................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TAAACTGACCTACGAGAGTCGCGCCA..... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................................TACGAGAGTCGCGCCAATCTG | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................GACCTACGAGAGTCGCGCCAATCTG | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................TAGGTCTCTGTGCAGGTTTT........................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TCGGAGCCAACGACTTGCAGC................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ATAGGTCTCTGTGCAGGTTTA........................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GATAGGTCTCTGTGCAGGTAT............................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ATAGGTCTCTGTGCAGGTTT............................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................GCCAACGACCTGTAGCG.................................. | 17 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ATAGGTCTCTGTGCAGGATTT........................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................TACCCAGATGAAAAGA............................................................................................................................................................................................................... | 16 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................TTTTGAAACTGAAGTG............................................................................... | 16 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................TAAGCTATTGAAGAAGAT................................................................................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................TAAGAAGCGGCGTTAAA................................................................................................................ | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................TAAGCTATTGAAGAAGA.................................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ATACCTTTTACTGTGACGGTTTAAGCTAAGGGTCTACTTTTCTCAATACATGAAACCCTAGGGGTTACACACTTCGATAACTTCTTTTAACCGTATCCATATTTTTCGTCTGTATATCGCTATTCTTCGCCGCTATTCTATCCAGAGGCACGTGCAAAATTTTGACTTCACAATTGATTAGTGGTAGTGAAAGGGGCGTCGGTTGCTGAACGTCGCGTGATTTGACTGGATGCTCTCAGCGCGGTTAGAC
**************************************************.....((((......(((.((((.((((...........))))..............(((......((((.......((((.(((.......))).))))...))))......))).(((....)))..))))))).....)))).....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
SRR902008 ovaries |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................ATATCCAGAGGCACGTGCAAAATTT........................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................GGATGCTCTCAGCGCGGTTAGAC | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CAGAGGCACGTGCAAAATTTT....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TCGCGTGATTTGGCTGGATGC................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................GGCACGTCCAAAAATTTG...................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TGAAGGGGGCCTCGGTT.............................................. | 17 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| ........................................................CCTAGCGCTTACACACT................................................................................................................................................................................. | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................GGTTGACTCGATGCTCTC............. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................................TGCTGAGCGTCGAGTG............................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/24/2015 at 05:47 AM