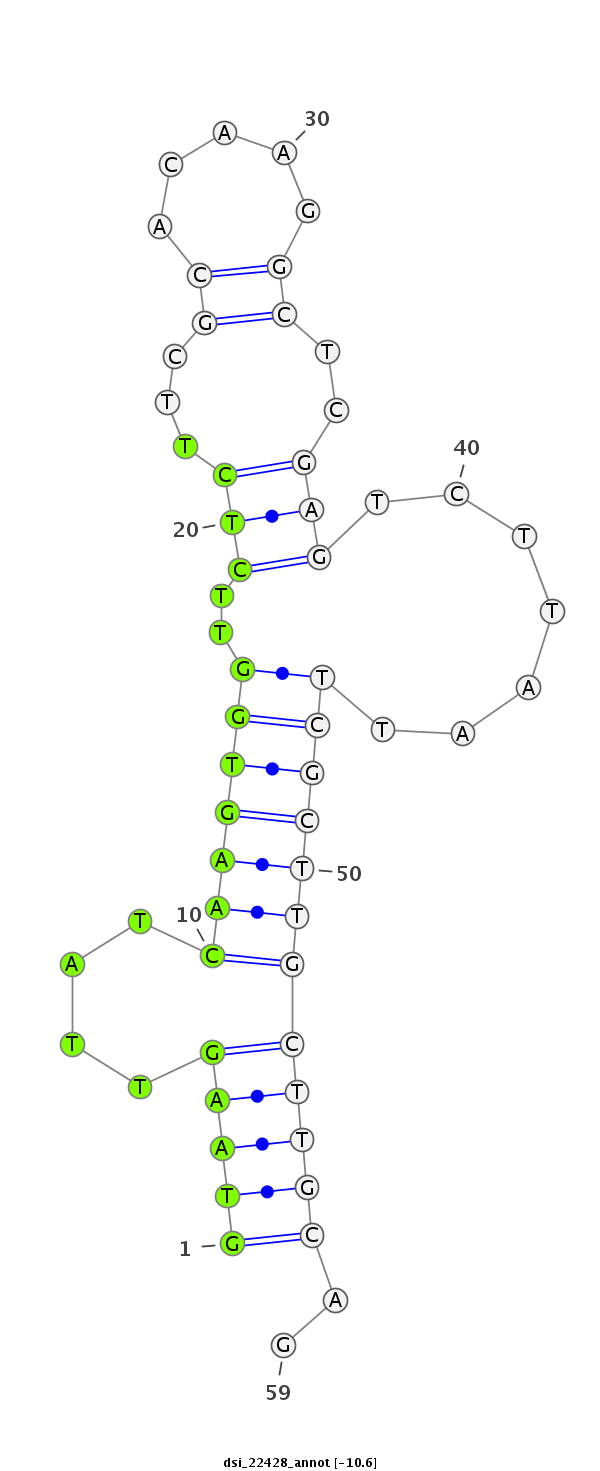
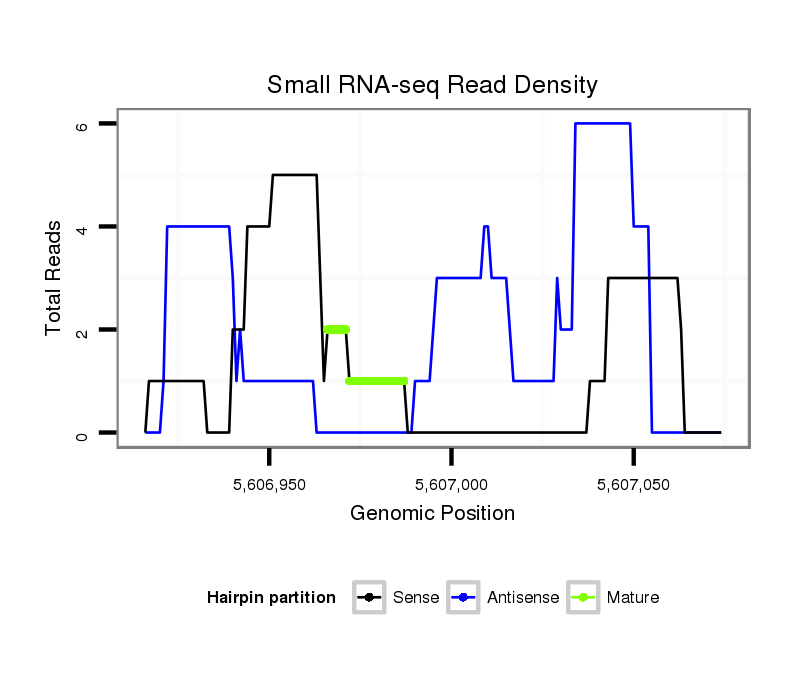
ID:dsi_22428 |
Coordinate:2r:5606966-5607024 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
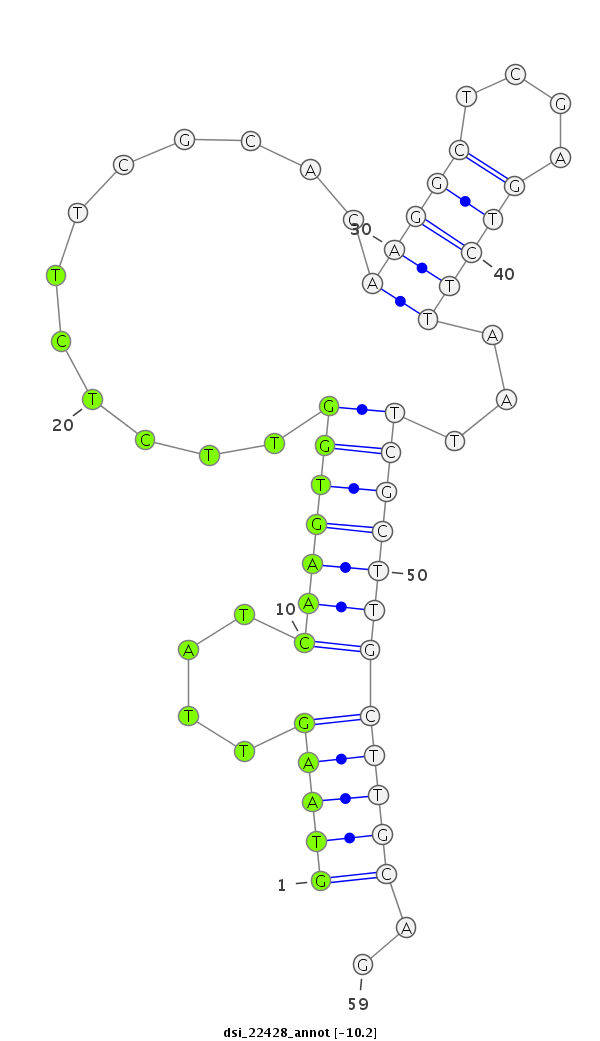
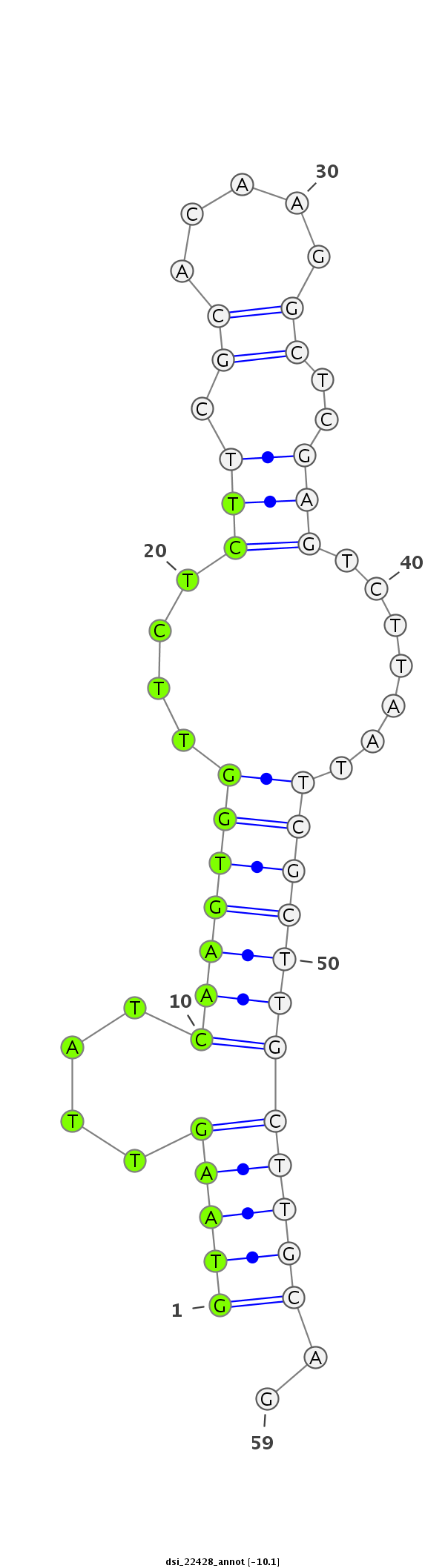
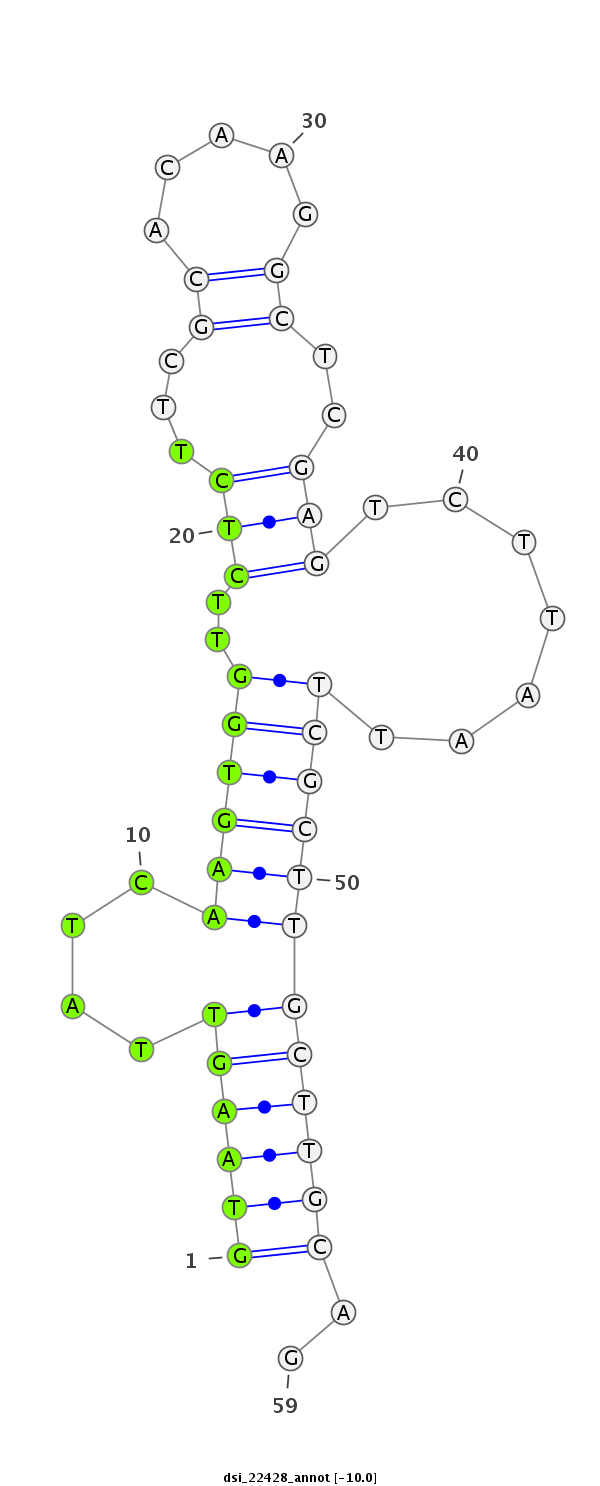
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -10.2 | -10.1 | -10.0 |
 |
 |
 |
exon [2r_5606868_5606965_+]; CDS [2r_5606868_5606965_+]; CDS [2r_5607025_5607188_+]; exon [2r_5607025_5607264_+]; intron [2r_5606966_5607024_+]
No Repeatable elements found
| ##################################################-----------------------------------------------------------################################################## GGTCAGTCATCTCAACGATAACAATGCCCTGATCCTGGACGAAGACCCAGGTAAGTTATCAAGTGGTTCTCTTCGCACAAGGCTCGAGTCTTAATTCGCTTGCTTGCAGATGAGCCCCAGTCCGAAGCAGAAAAGAACTGGAGCGATCTGGAGGAGGAT **************************************************(((((....(((((((..(((...((.....))..))).......))))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902008 ovaries |
SRR902009 testis |
M023 head |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................CAGAAAAGAACTGGAGCGATC........... | 21 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................CGGAAGCAGAAAAGAACTGGAGCGATC........... | 27 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................GGAGCGATCTGGAGGAGGT. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................CGAAGCAGAAAAGAACTGGAGCGAT............ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GTAAGTTATCAAGTGGTTCTCT....................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................CTGATCCTGGACGAAGACCC............................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .GTCAGTCATCTCAACG.............................................................................................................................................. | 16 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................TGCCCTGATCCTGGACGAAGACCC............................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................TGGACGAAGACCCAGGTAAGT....................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................TGCCCTGATCCTGGACGAAGACCCA.............................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................CTGATCCTGGACGAAGACCCA.............................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................CCTGATCCTGGACGAAGACCCAGA............................................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................CCCTGATCCTGGACGAAGACCCAGA............................................................................................................ | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................CCTGATCCGGGTTGAAGAC................................................................................................................. | 19 | 3 | 13 | 0.69 | 9 | 0 | 0 | 0 | 0 | 6 | 3 | 0 | 0 |
| ....................................................................................................................................AAGAACTGGAGCGATTGCGAG...... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................CTGATCCTGGTTGAAGAC................................................................................................................. | 18 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................GCTGATCCGGGTCGAAGAC................................................................................................................. | 19 | 3 | 14 | 0.21 | 3 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 |
| ............................CTGATCCGGGTCGAAGAC................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................TGATCCGGGTCGAAGAC................................................................................................................. | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................GATCCCGGACGAACCCCCA.............................................................................................................. | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................TCTGGAGCGGTCTGGAGCA.... | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................TGATCCTGGTTGAAGACA................................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................CCTGGACGAAGACTGACG............................................................................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CCAGTCAGTAGAGTTGCTATTGTTACGGGACTAGGACCTGCTTCTGGGTCCATTCAATAGTTCACCAAGAGAAGCGTGTTCCGAGCTCAGAATTAAGCGAACGAACGTCTACTCGGGGTCAGGCTTCGTCTTTTCTTGACCTCGCTAGACCTCCTCCTA
**************************************************(((((....(((((((..(((...((.....))..))).......))))))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
M025 embryo |
SRR902009 testis |
GSM343915 embryo |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................TCAGGCTTCGTCTTTTCTTGA.................... | 21 | 0 | 1 | 4.00 | 4 | 1 | 2 | 1 | 0 | 0 | 0 |
| .................................................................................................................CGGGGTCAGGCTTCGTCTTTT......................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CCGAGCTCAGAATTAAGCGAA.......................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................TCCGAGCTCAGAATTAAGCGA........................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......AGTAGAGTTGCTATTGTTA...................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......AGTAGAGTTGCTATTGTTACG.................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......AGTAGAGTTGCTATTGTT....................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................CGTGTTCCGAGCTCAGAATTA................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....CAGTAGAGTTGCTATTGTTA...................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................GGGACTAGGACCTGCTTCTGG................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................GGCGTGTTCCGAGCTCAGAAT.................................................................. | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................TAAGCGAACGAACGTCTACTC............................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................GGTCCCTTCATTAGTTCAGC............................................................................................. | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................GCGGGGTCAGTCTTCG............................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................TTTGCGGGACTAGGAGCTG....................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
Generated: 04/23/2015 at 06:52 PM