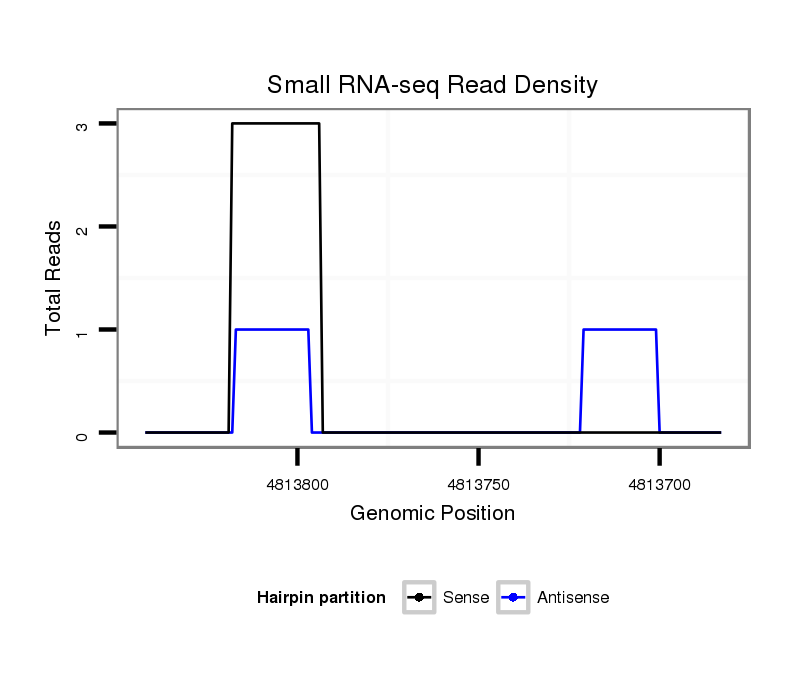
ID:dsi_22411 |
Coordinate:2r:4813733-4813792 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_4813580_4813732_-]; CDS [2r_4813580_4813732_-]; CDS [2r_4813793_4814071_-]; exon [2r_4813793_4814071_-]; intron [2r_4813733_4813792_-]
No Repeatable elements found
| ##################################################------------------------------------------------------------################################################## CAAACAAATTGTTTAGAATCAAGATCAACCAAATACAGATGAAGGAGACAGTAAGTGCCTTTAGTTTTGATGTAATTTGCACAGACCTAATCTTTCCCCTTCTCCATTAGAATGAGGAGCAAAAGGCGACGGAGGAGCGTGTATTCCAGGATCGCCAGTA **************************************************..........((((.((((.((((...)))))))).))))....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ........................TCAACCAAATACAGATGAAGGAGAC............................................................................................................... | 25 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| ...........................................................................................................TGGAATGAGGAGCAAAAGGCGA............................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ................................................................TTTTGATCTAGTTTGCACAGA........................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...............................................................................................................CTGAGGAGCAAAAGG.................................. | 15 | 1 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ................................................................................................................TGAGGAGCAAAAGGA................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
GTTTGTTTAACAAATCTTAGTTCTAGTTGGTTTATGTCTACTTCCTCTGTCATTCACGGAAATCAAAACTACATTAAACGTGTCTGGATTAGAAAGGGGAAGAGGTAATCTTACTCCTCGTTTTCCGCTGCCTCCTCGCACATAAGGTCCTAGCGGTCAT
**************************************************..........((((.((((.((((...)))))))).))))....................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
M025 embryo |
O001 Testis |
M053 female body |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR1275487 Male larvae |
GSM343915 embryo |
SRR553485 Chicharo_3 day-old ovaries |
SRR902008 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................CAGACTACTCGCACATAAGG............. | 20 | 3 | 2 | 561.50 | 1123 | 632 | 449 | 29 | 6 | 3 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................CAGACTACTCGCACATAAG.............. | 19 | 3 | 13 | 56.38 | 733 | 399 | 313 | 11 | 1 | 2 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GACTACTCGCACATAAGG............. | 18 | 2 | 2 | 3.50 | 7 | 0 | 0 | 0 | 6 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCAGACTACTCGCACATAAGG............. | 21 | 3 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CAGACTCCTCGCACATAAG.............. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................TTTCCGCTGCCTCCTCGCACA.................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CAGACTACTCGCACATAAGGTC........... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................ATGTCTTCTGCCTGTGTCAT........................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GACTACTCGCACATAAGGT............ | 19 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................GTTGGTTTATGTCTACTTCCT.................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......TACCAAATCTTAGTTCTA....................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCAGACTACTCGCACATAA............... | 19 | 3 | 9 | 0.78 | 7 | 5 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................CAGTCTACTCGCACATAAGG............. | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GTTTTTGTAACAAAACTTAGTTC......................................................................................................................................... | 23 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................GACTACTCGCACATAAGGCC........... | 20 | 3 | 7 | 0.43 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................CTCGCACATAAGGCCCTG........ | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................GTTCTAGTTGGTCTAT............................................................................................................................. | 16 | 1 | 7 | 0.29 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................GCAGACTACTCGCACATAAG.............. | 20 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTCGCACATAAGGCC........... | 15 | 1 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................AGGTCGTAGCGGTCA. | 15 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTCGTACATAAGGCCCTGG....... | 19 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................TCGTTCTAGTTGGTCTAT............................................................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........CAACTCCTAGTTCTAGATGG.................................................................................................................................. | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................TCCTAGTTGGTTTAT............................................................................................................................. | 15 | 1 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................CAGGCTACTCGCACATAAG.............. | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............GATCTTAGTTCGAGTTG................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................TTAGTTCTAGTTCGT................................................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................ATCGTTCTAGTTGGTCTAT............................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............TTTTAGTTCTAGTTGGC................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/23/2015 at 05:38 AM