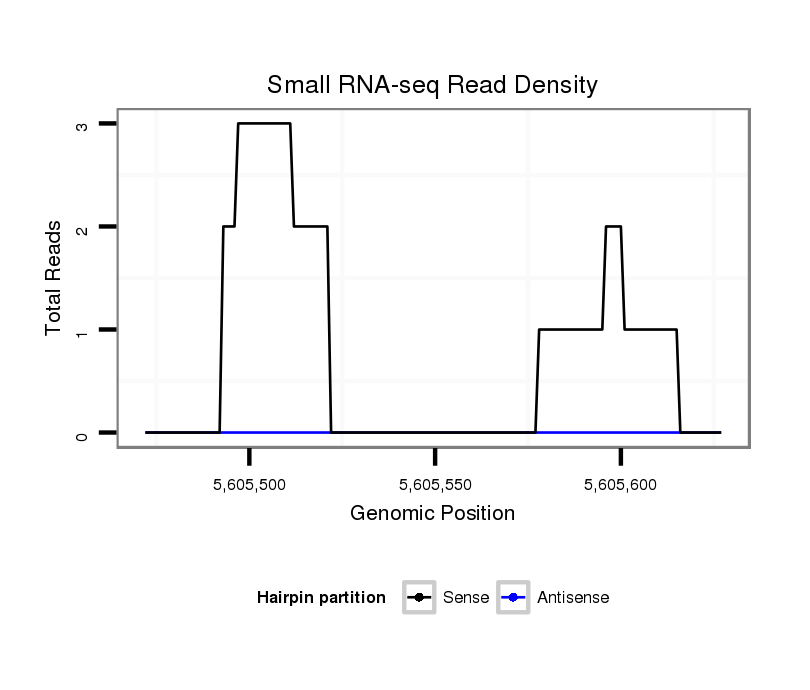
ID:dsi_21998 |
Coordinate:2r:5605522-5605577 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
exon [2r_5605393_5605521_+]; CDS [2r_5605393_5605521_+]; CDS [2r_5605578_5605727_+]; exon [2r_5605578_5605727_+]; intron [2r_5605522_5605577_+]
No Repeatable elements found
| ##################################################--------------------------------------------------------################################################## AATCCCTTGATTAACGAAATTTCCATCCTGCTGCAAGGAAAGCACGCGAGGTAAGTGTTTTCCGTAGAGCCCTATTGAGTCTTTTTCTAAAGAAGTTATTTTACAGTGTTCGCTCCGCCCTGCGTTGGCTGCACGTCCTGACCTCCACCCACACTA **************************************************(((((((.((((..(((((...............)))))..)))).)).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M023 head |
O001 Testis |
|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................TGTTCGCTCCGCCCTGCGTTGGC........................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................TCCTGCTGCAAGGAAAGCACGCGAG.......................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................TTGGCTGCACGTCCTGACCT............ | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................TCCATCCTGCTGCAAGGAAAGCACGCGAG.......................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................TCCATCCTGCTGCAAGGAA.................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............GAAATTTCGATCCTG.............................................................................................................................. | 15 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................GTAGAGCACTGTTTAGTCT.......................................................................... | 19 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......TTGATTAACGAGATT....................................................................................................................................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......TGATTAACGAAATCT...................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
|
TTAGGGAACTAATTGCTTTAAAGGTAGGACGACGTTCCTTTCGTGCGCTCCATTCACAAAAGGCATCTCGGGATAACTCAGAAAAAGATTTCTTCAATAAAATGTCACAAGCGAGGCGGGACGCAACCGACGTGCAGGACTGGAGGTGGGTGTGAT
**************************************************(((((((.((((..(((((...............)))))..)))).)).)))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TTACGTGCAGGACTGGAGGTGG....... | 22 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........TTTGCTTTAAAGGTAGGACGACG.......................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AATGTCACAAGCCTGG........................................ | 16 | 2 | 14 | 0.71 | 10 | 0 | 10 | 0 | 0 | 0 | 0 |
| .........AAATTGCTTTGAAGGTAGG................................................................................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........ATTGATCTAGAGGTAGGACG............................................................................................................................. | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................TCAGAAGAAGATTTCT............................................................... | 16 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................ACAAAGGGCATCTCGTG.................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................GCAGGACTGGAGGTGCGGA.... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................CGTGCAGGCCTGGCGATG........ | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/23/2015 at 06:41 PM