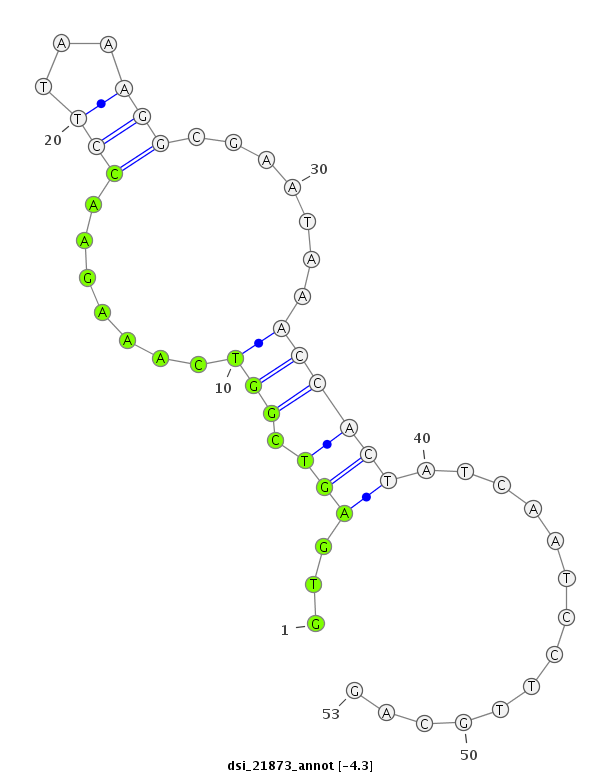
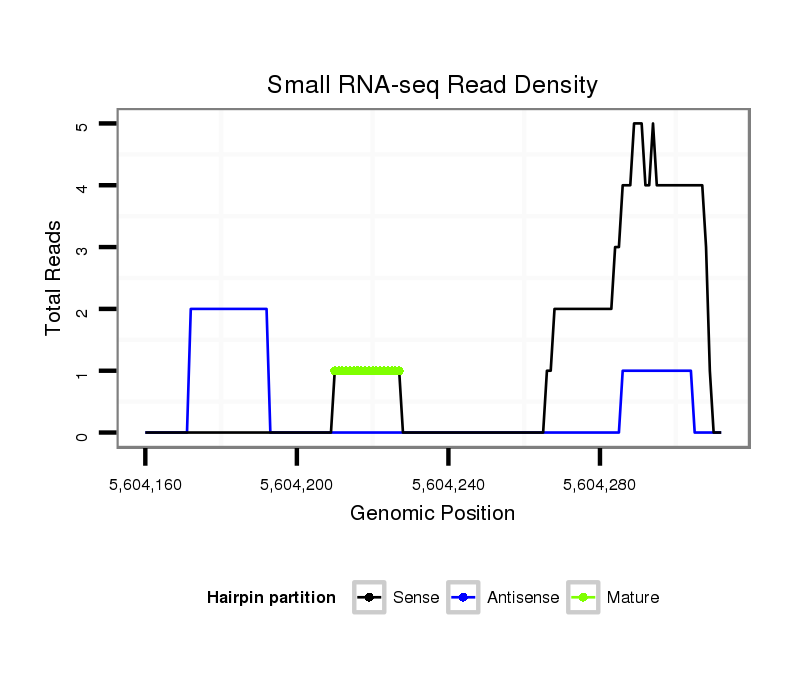
ID:dsi_21873 |
Coordinate:2r:5604210-5604262 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
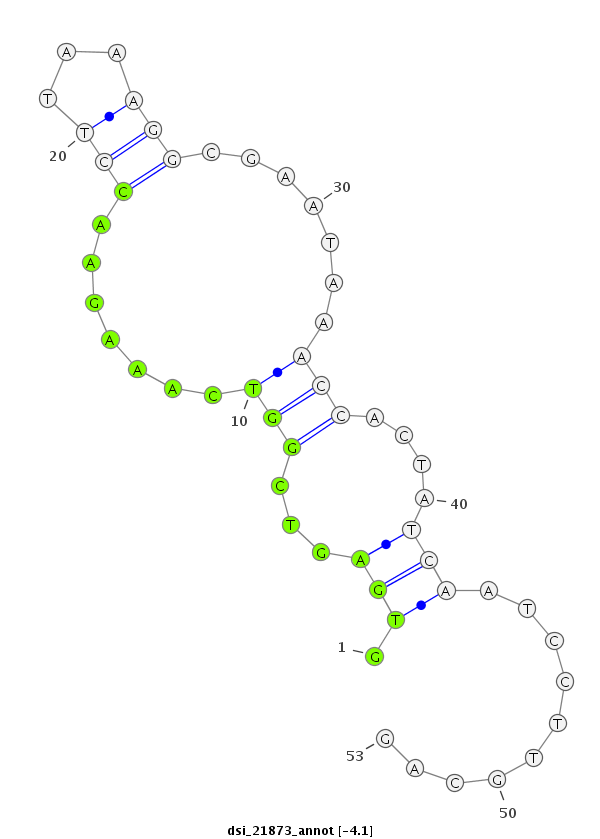
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -4.1 | -3.7 | -3.4 |
 |
 |
 |
CDS [2r_5603907_5604209_+]; exon [2r_5603836_5604209_+]; CDS [2r_5604263_5604792_+]; exon [2r_5604263_5604792_+]; intron [2r_5604210_5604262_+]
No Repeatable elements found
| ##################################################-----------------------------------------------------################################################## TGGGAACGTACAAGGGCTCAGTGGTTATATACTCATTGGCCGAGGGAAAGGTGAGTCGGTCAAAGAACCTTAAAGGCGAATAAACCACTATCAATCCTTGCAGATTTCTCGAACACTCAGTGGTGACAGCCATGATGGACCAGTGACCTCAAT **************************************************...(((.(((.......(((...))).......))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
GSM343915 embryo |
M024 male body |
M023 head |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................CCATGATGGACCAGTGACC..... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................GAAAGATTTCTCGAACACTCAGT................................ | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................GTGAGTCGGTCAAAGAAC..................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TCGAACACTCAGTGGTGACAGCCATGA.................. | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................GACAGCCATGATGGACCAGTGACCTC... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................CAGCCATGATGGACCAGTGACCT.... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................TATATACTCATTGGCCGAGGGAAAGA...................................................................................................... | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TCTCGAACACTCAGTGGTGACAGCCA..................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................ATGGACCAGTGACCT.... | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................................................CTAGTGACAGCCATGAT................. | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................CCGGGGGAAAGGTGA................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................GGACAGCCATGATGGAAGAG.......... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................GAGACGGTCATGATGGAC............. | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
|
ACCCTTGCATGTTCCCGAGTCACCAATATATGAGTAACCGGCTCCCTTTCCACTCAGCCAGTTTCTTGGAATTTCCGCTTATTTGGTGATAGTTAGGAACGTCTAAAGAGCTTGTGAGTCACCACTGTCGGTACTACCTGGTCACTGGAGTTA
**************************************************...(((.(((.......(((...))).......))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ............TCCCGAGTCACCAATATATGA........................................................................................................................ | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................GTCGGTACTACCTGGTCAC........ | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................TCCCCACTGTCAGCACTAC................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ............ACCCGAGTCACCAATCT............................................................................................................................ | 17 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| .............CCCGAGTCACCAATC............................................................................................................................. | 15 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................AATAGTTAGGAACGT................................................... | 15 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 06:35 PM