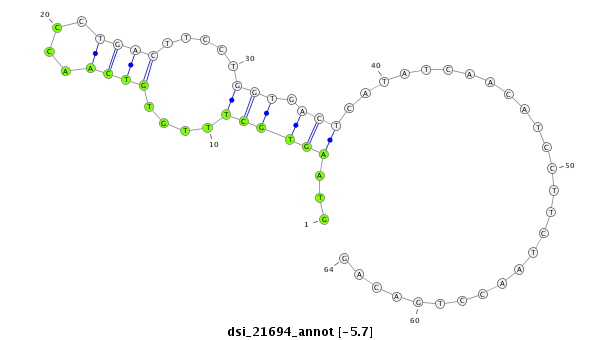
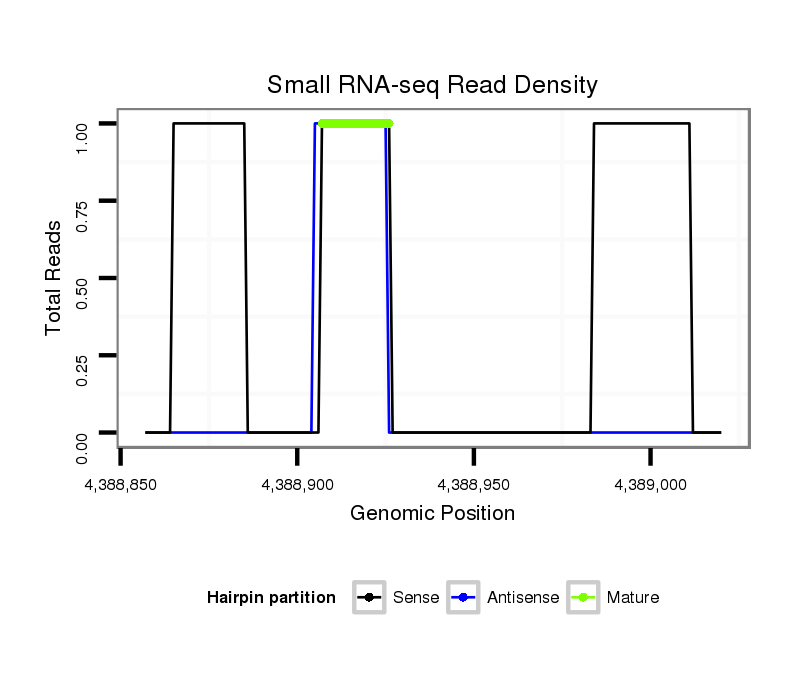
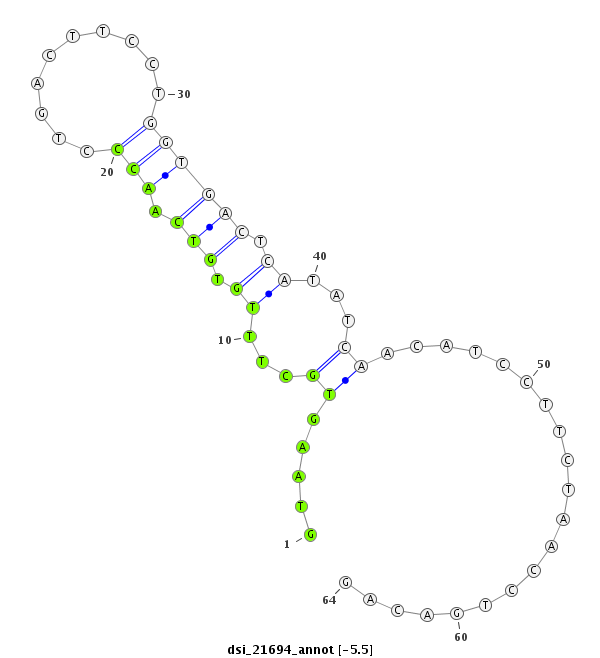
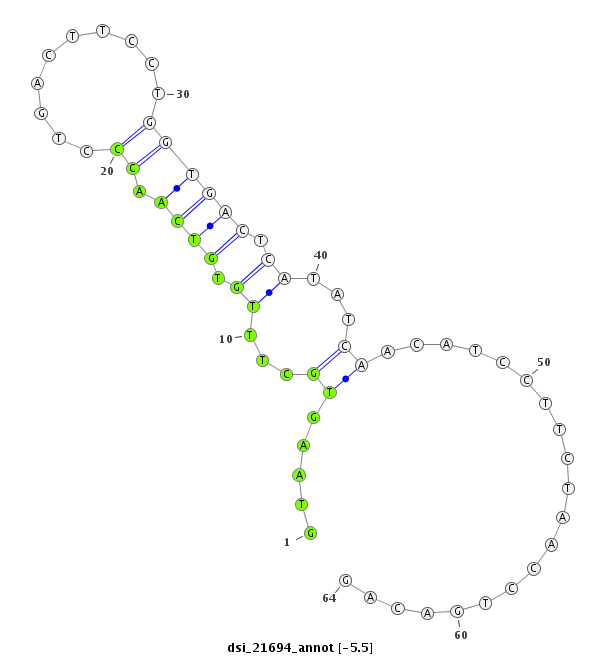
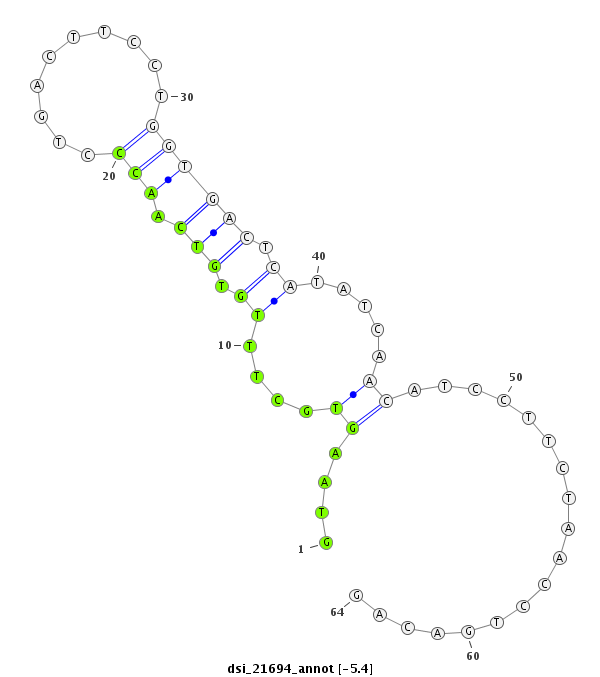
ID:dsi_21694 |
Coordinate:3r:4388907-4388970 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -5.5 | -5.5 | -5.4 |
 |
 |
 |
exon [3r_4388827_4388906_+]; CDS [3r_4388827_4388906_+]; CDS [3r_4388971_4389142_+]; exon [3r_4388971_4389142_+]; intron [3r_4388907_4388970_+]
No Repeatable elements found
| ##################################################----------------------------------------------------------------################################################## GGACATGCAGCAGCAGCATCCGCAGGGCTATGCCTCCAGGCGTTCGCAAGGTAAGTGCTTTGTGTCAACCCTGACTTCCTGGTGACTCATATCAACATCCTTCTAACCTGACAGACTCCTGCAATCAGTATAAGGAATCGGGCAAGTCAAGCGACTCTAGTCTG **************************************************...((((((....((((....)))).....))).)))...........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................GTATAAGGAATCGGGCAAGTCAAGCGAC......... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ........AGCAGCAGCATCCGCAGGGCT....................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................GTAAGTGCTTTGTGTCAACC.............................................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................ACTTCCTGGGGACTGATATAA...................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................CTTCCTGGGGACTGATATAA...................................................................... | 20 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ....................................CTGGCGTTCGCAAGG................................................................................................................. | 15 | 1 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 |
| ..........................................................................CTTCCTGGGGACTGATATTA...................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................GACTTCCTGGAGACT............................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| ...................................................TAAGTGCTTTGTTTGAA................................................................................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
CCTGTACGTCGTCGTCGTAGGCGTCCCGATACGGAGGTCCGCAAGCGTTCCATTCACGAAACACAGTTGGGACTGAAGGACCACTGAGTATAGTTGTAGGAAGATTGGACTGTCTGAGGACGTTAGTCATATTCCTTAGCCCGTTCAGTTCGCTGAGATCAGAC
**************************************************...((((((....((((....)))).....))).)))...........................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
O001 Testis |
SRR553487 NRT_0-2 hours eggs |
M023 head |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................GTAGGAAAATTGGAAGGTC.................................................. | 19 | 3 | 20 | 1.15 | 23 | 0 | 0 | 0 | 11 | 11 | 0 | 0 | 1 |
| ................................................TCCATTCACGAAACACAGTTG............................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........CGCAGTCGTAGGCGTCCC......................................................................................................................................... | 18 | 2 | 6 | 0.83 | 5 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................AGAATGGACTCTCTGAGGAT........................................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TGTCTGAGGAGGTTTGTCA................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............AGTCGTAGGCGTCCC......................................................................................................................................... | 15 | 1 | 19 | 0.26 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GGTATAGTTGTAAGAAGA............................................................ | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ................................................................GGTTGGGACTGATGGAC................................................................................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................AAAGTTGGGACTGAA....................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TAGAGTTGTAGGACGAT........................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................TTGGACTGTCACAGGA............................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
Generated: 04/24/2015 at 05:07 AM