ID:dsi_2156 |
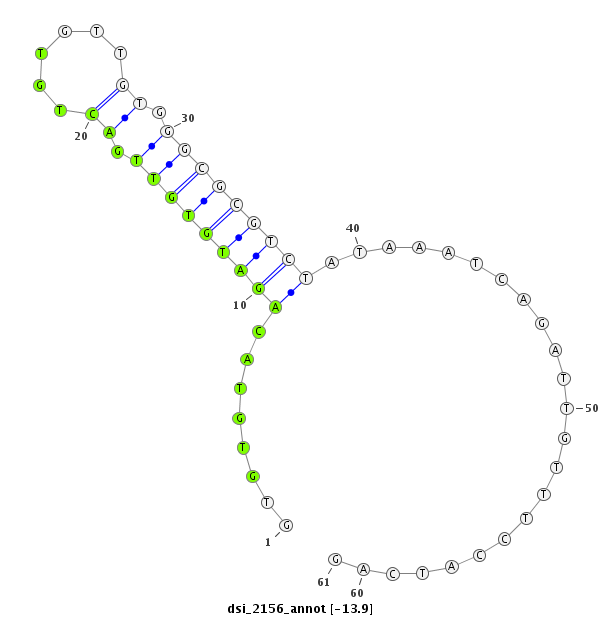
Coordinate:2r:5846230-5846290 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
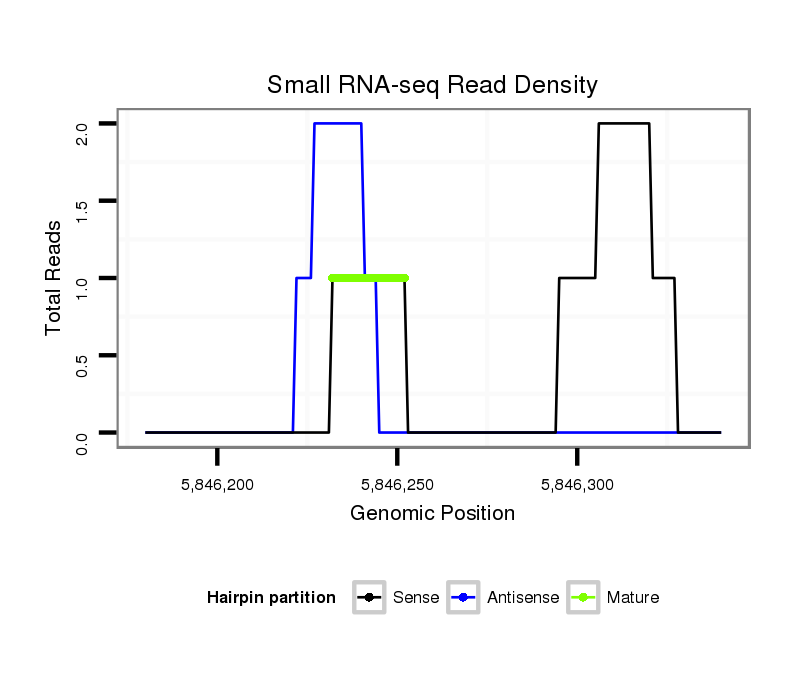
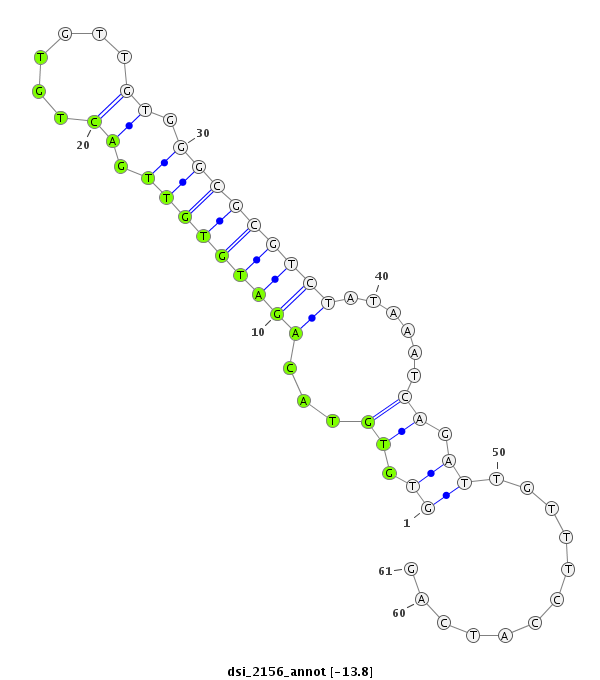
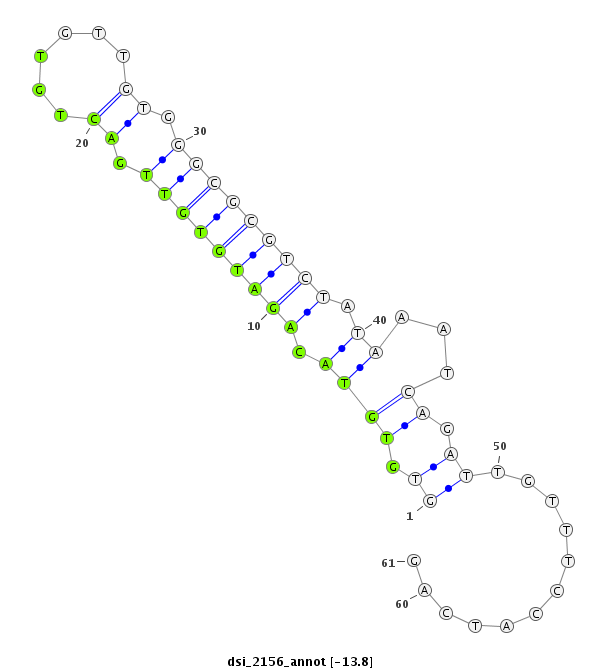
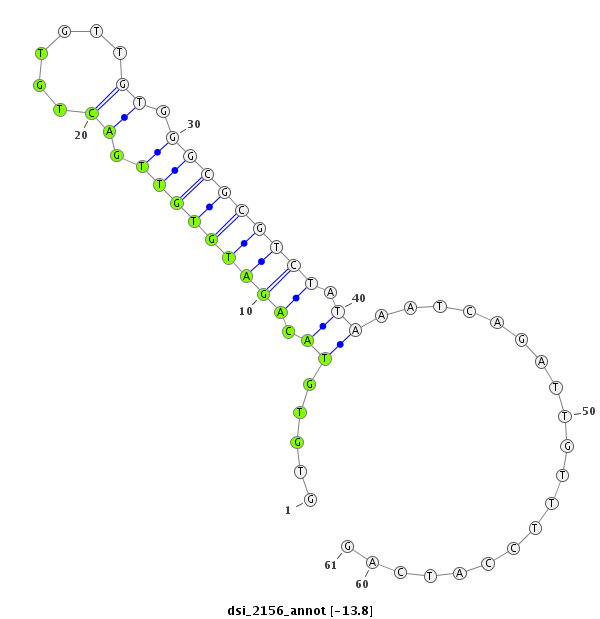
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -13.8 | -13.8 | -13.8 |
 |
 |
 |
CDS [2r_5846058_5846229_+]; exon [2r_5845891_5846229_+]; CDS [2r_5846291_5846403_+]; exon [2r_5846291_5846403_+]; intron [2r_5846230_5846290_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------################################################## TATTTTAGAGCCCGCCGTATTTCTGGTTTTCTTTGGCAGATTCCTAACAGGTGTGTACAGATGTGTTGACTGTGTTGTGGGCGCGTCTATAAATCAGATTGTTTCCATCAGATGCCGTTTACCAGAACCAGATACTCTACCAGACCTGCGTCACGGTCATG **************************************************........(((((((((.((......)).))))))))).......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................GTGTACAGATGTGTTGACTGT........................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................CGTTTACCAGAACCAGATACTCTACC.................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................ACCAGATACTCTACCAGACCTG............. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................TGCCGTTTACCAGACCCGA.............................. | 19 | 3 | 13 | 0.15 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ................................................................................................................TGCCGGTTACCAGAACCGA.............................. | 19 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................CCAGAATCCTAACAGG.............................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................................GCTGTGTACGGAGGTGTTG............................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
ATAAAATCTCGGGCGGCATAAAGACCAAAAGAAACCGTCTAAGGATTGTCCACACATGTCTACACAACTGACACAACACCCGCGCAGATATTTAGTCTAACAAAGGTAGTCTACGGCAAATGGTCTTGGTCTATGAGATGGTCTGGACGCAGTGCCAGTAC
**************************************************........(((((((((.((......)).))))))))).......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TTGGTCTACGCGATGGTCTG................ | 20 | 2 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GGGTTTGGTCTACGAGATGGTCTG................ | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGTCTACGCGATGGTCTG................ | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................GGATTGTCCACACATGTCT.................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TTGGTCTATGCGATGGT................... | 17 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................GTCCACACATGTCTACAC................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TTGGTCTACGCGATGGTCTGA............... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................TTGCTCCATGAGATGGT................... | 17 | 2 | 10 | 0.40 | 4 | 4 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................TATGAGATGGTCAGG............... | 15 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGGTCTACGCGATGGTCTGA............... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................TATGAGATGGACTGGT.............. | 16 | 2 | 20 | 0.25 | 5 | 4 | 0 | 1 | 0 | 0 |
| .............................................................................................................................TTGCTCCATGAGATGG.................... | 16 | 2 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 |
| .............................................................................................AGTCTTACTAAGGTAGTATA................................................ | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................AAATGGTCTTGGTCTTTT.......................... | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................AGGGTTGTCCACACA......................................................................................................... | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................TTGGTCTGCGAGATGGT................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................CGGCAAATGGTCGTGG................................ | 16 | 1 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................GGTCTACGCGATGGTCTGA............... | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGAGTCTAACAAAGGTAAT................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..AAAATCTCGGGTTGCAT.............................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................GGCAAATGGTCTTGA................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
Generated: 04/24/2015 at 01:14 AM