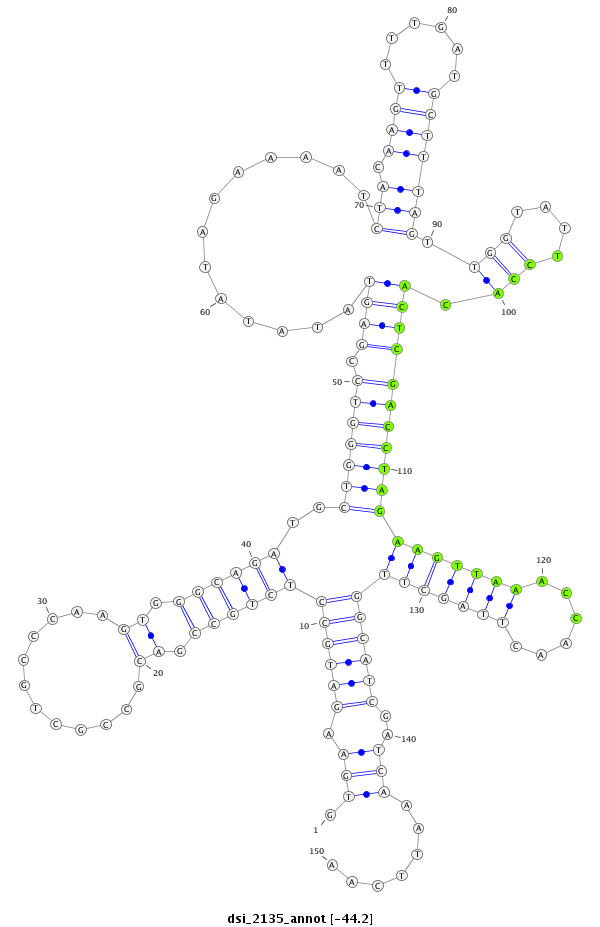
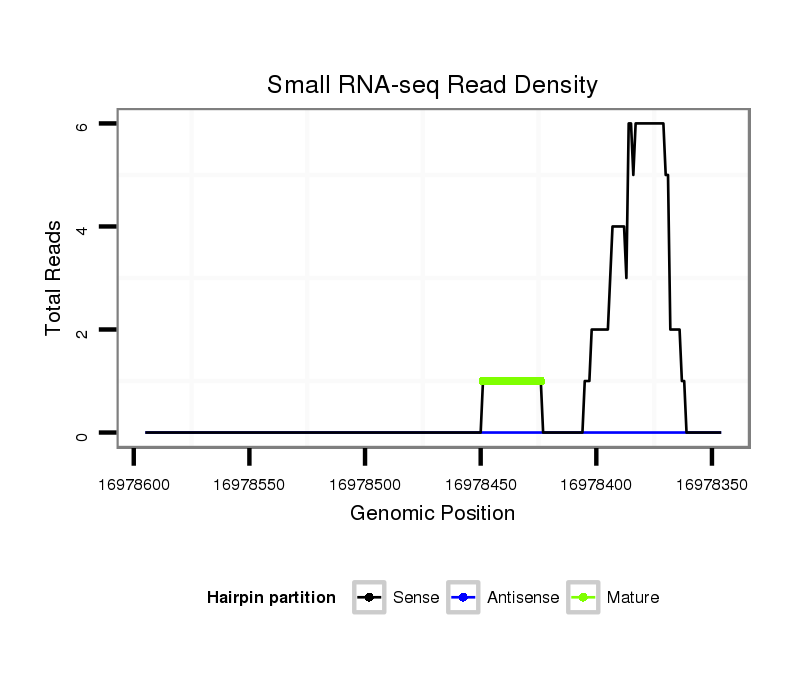
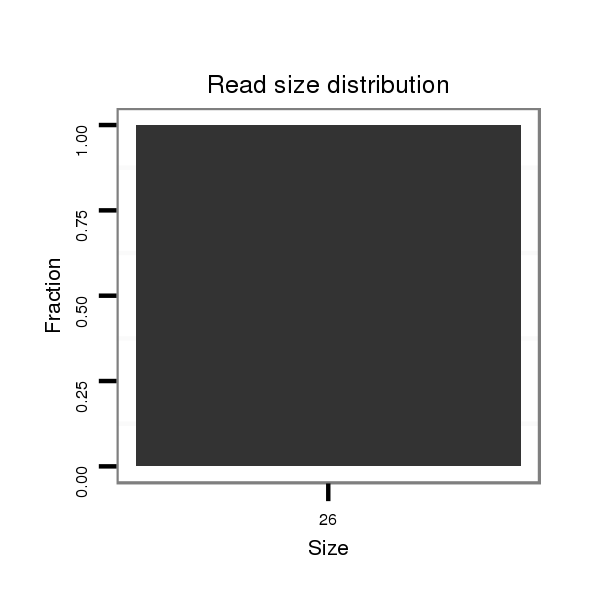
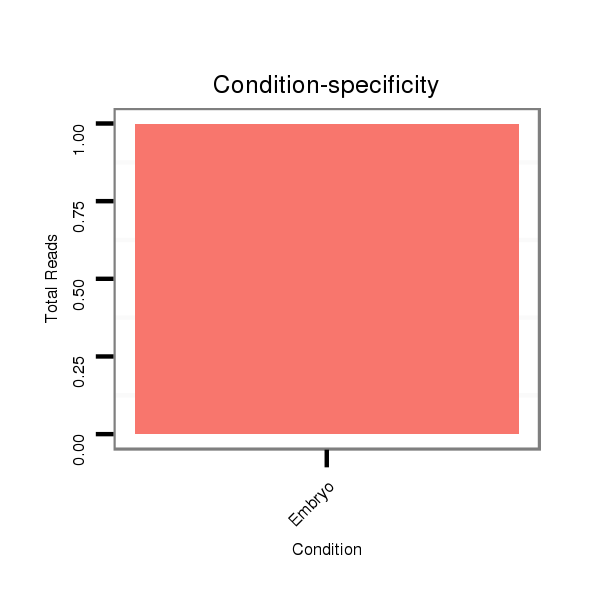
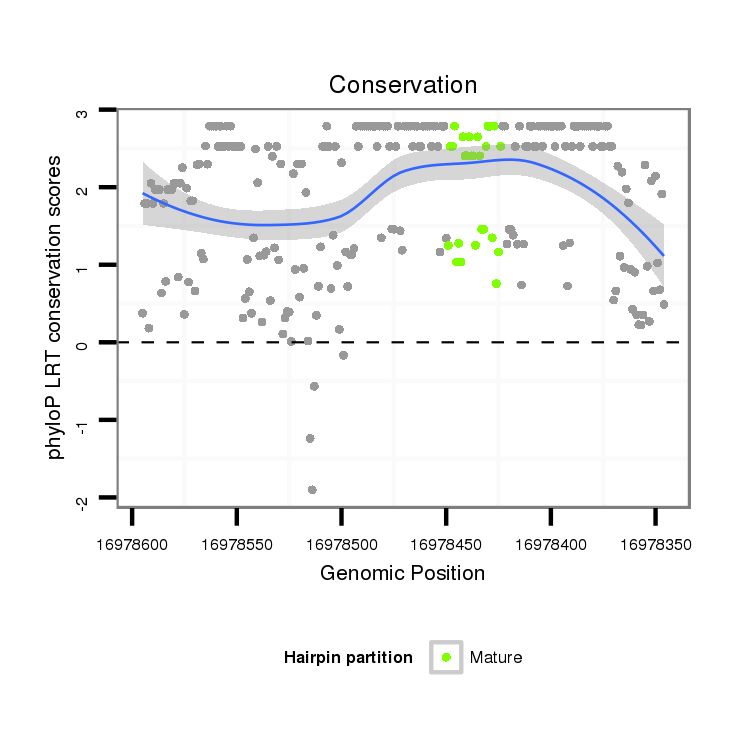
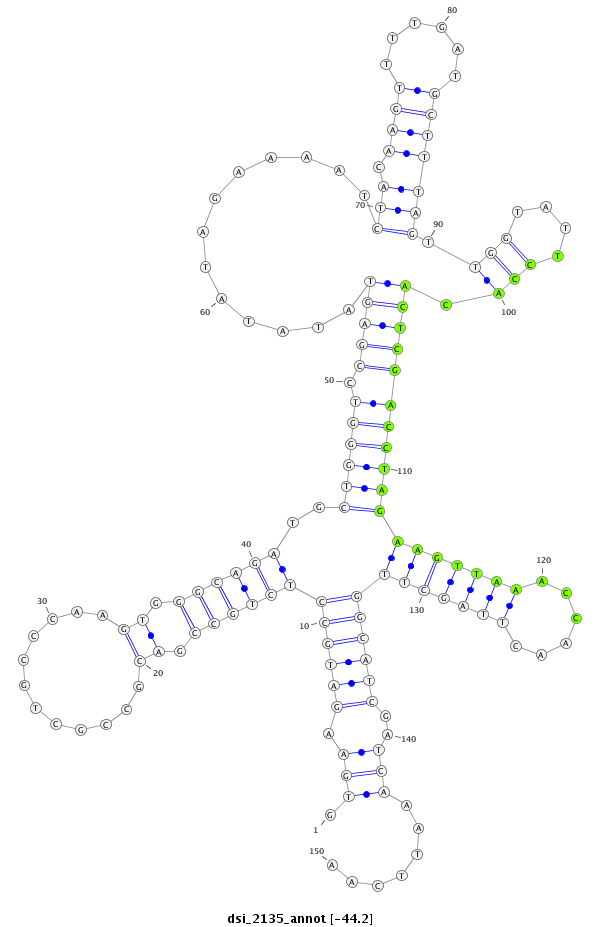
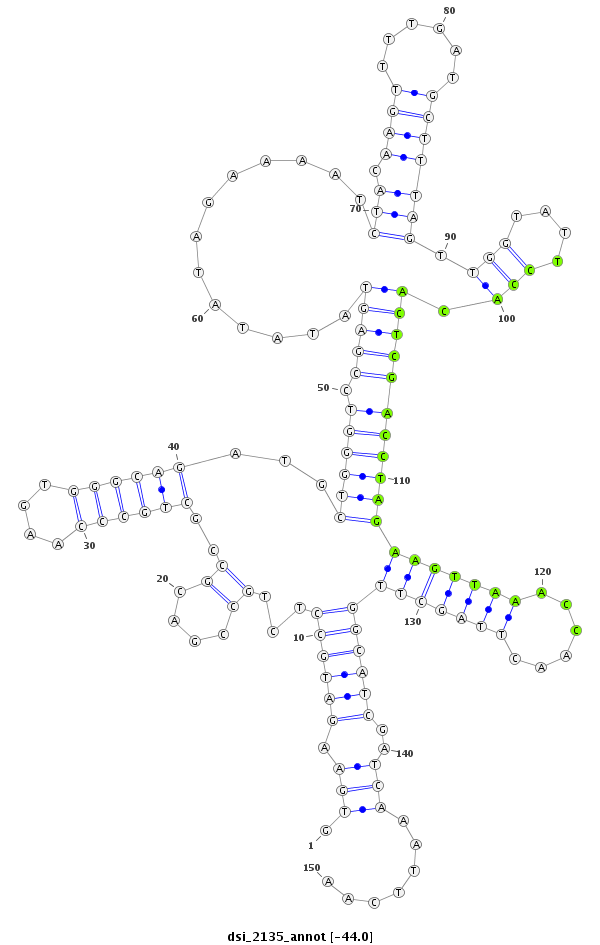
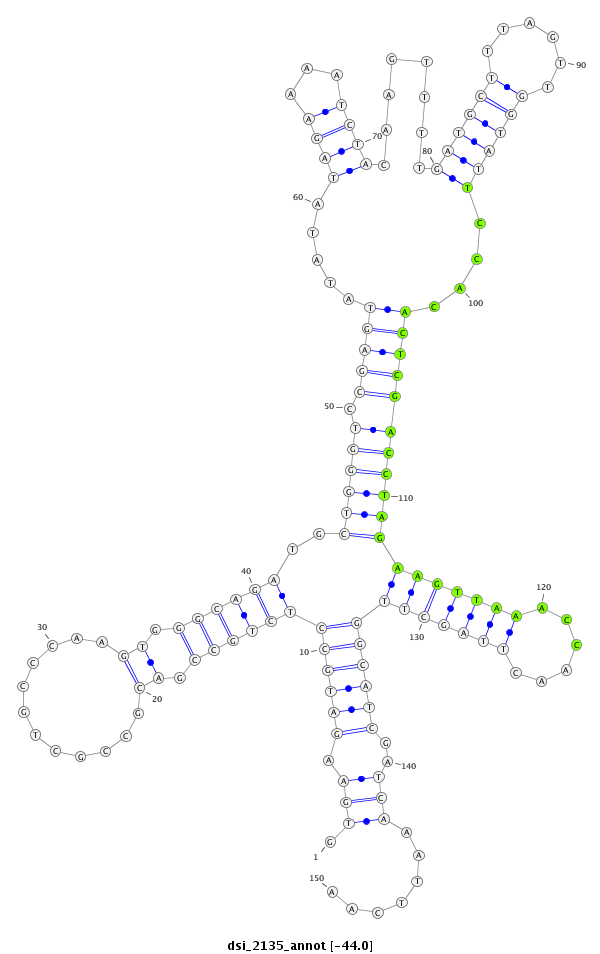
| AAA-----CGCTTTCGATT-TCGGC------------------------TT-TTG--GCC---------------------------------G------TGT-GTTAACAGCTGTCGCCCAGGTGAAG---ATG--------CC--------TCTGCC------------G------------ACGCCGCTGCCCAG----------------------------GTG----GGCAGATGC------------------------TG----------------------------GG-----TCCGAGTATATAT----AGAAAATCTACAAGTTTTGATGCTTTAGTTGGTATTCCACACTCGACCTAGAAGTT-----AAACCAACT-----T---AGCTT-GGCATCGATC--AAATTCAAT-CGTTC--ATTGAAT-TTGATACGATCGGTCGCGTCTTCTCC---T-TTC----CGATCCT- | Size | Hit Count | Total Norm | Total | M055
Female-body | M057
Embryo | V060
Head | V040
Embryo | V107
Male-body | V108
Head | GSM1528801
follicle cells |
| .............................................................................................................................................................................................................................................................................................................................................................................................AAACCAACT-----T---AGCTT-GGCATC............................................................................. | 21 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................T---AGCTT-GGCATCGATC--AAA.................................................................... | 19 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................ATTCCACACTCGACCTAGAAGTT-----AAAC....................................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................ACCAACT-----T---AGCTT-GGCATCGATC--AAATTC................................................................. | 29 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................TC--ATTGAAT-TTGATACGATC................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................ATTCCACACTCGACCTAGAA................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................CTCGACCTAGAAGTT-----AAACCAACT-----T---......................................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................T-----AAACCAACT-----T---AGCTT-GGCATCGAT.......................................................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................ATTGAAT-TTGATACGATCGGTCGC............................. | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................AAACCAACT-----T---AGCTT-GGCATCGATC--A...................................................................... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................T-TTGATACGATCGGTCGCGT........................... | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................AACT-----T---AGCTT-GGCATCGATC--AAATT.................................................................. | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................CAACT-----T---AGCTT-GGCATCGATC--....................................................................... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................ACT-----T---AGCTT-GGCATCGATC--AA..................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................CGACCTAGAAGTT-----AAACCAAC................................................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................TGGTATTCCACACTCGACCTAGAA................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................CGACCTAGAAGTT-----AAACCAACT-----T---AGCT..................................................................................... | 27 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................AAT-CGTTC--ATTGAAT-TTGATACGAC.................................... | 25 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................GATCGGTCGCGTCTTCTCC---T-TTC----CGAT.... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................AAT-TTGATACGATCGGTCGCGT........................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................T-----T---AGCTT-GGCATCGATC--AAA.................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................T-----AAACCAACT-----T---AGCTT-GGCATC............................................................................. | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................AACT-----T---AGCTT-GGCATCGATC--AAA.................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................GATCGGTCGCGTCTTCTCC---T-TTC----C....... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................TATTCCACACTCGACCTAGAAG.................................................................................................................. | 22 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................CGCGTCTTCTCC---T-TTC----CGATC... | 21 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................C--ATTGAAT-TTGATACGATCGGTC............................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |