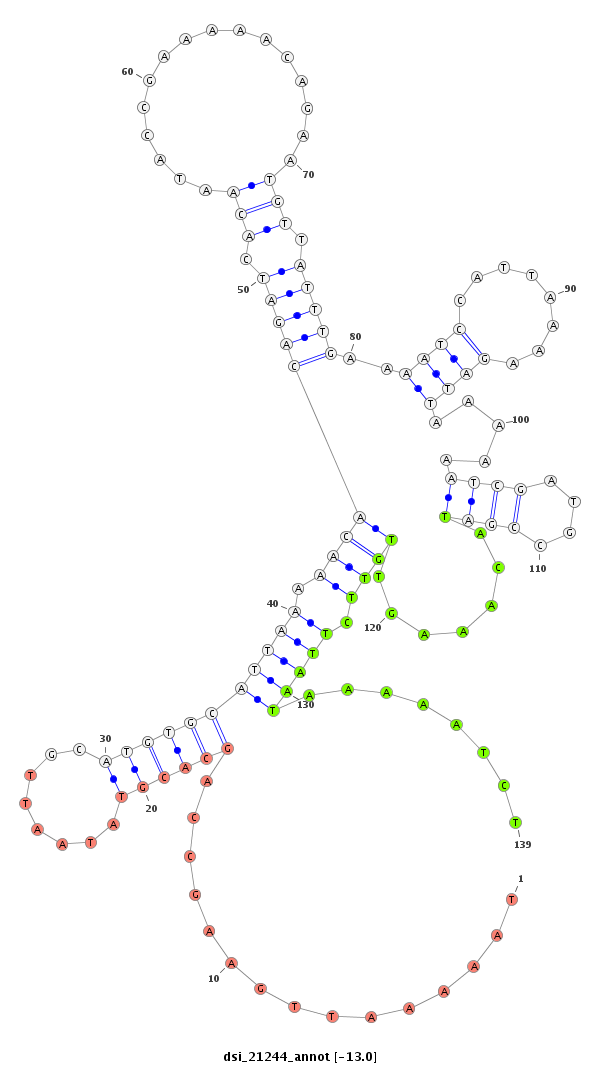
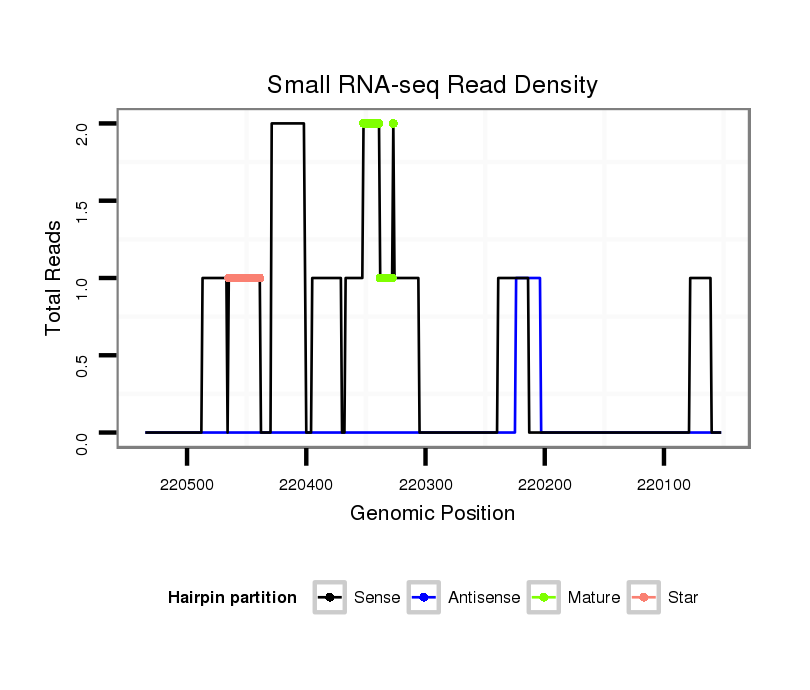
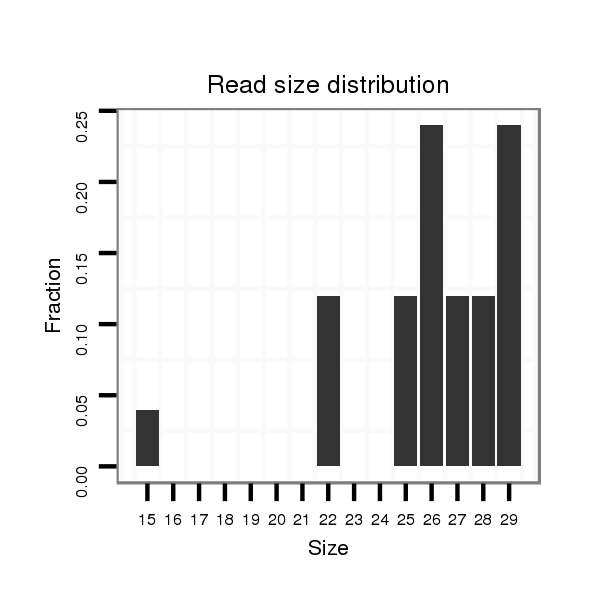
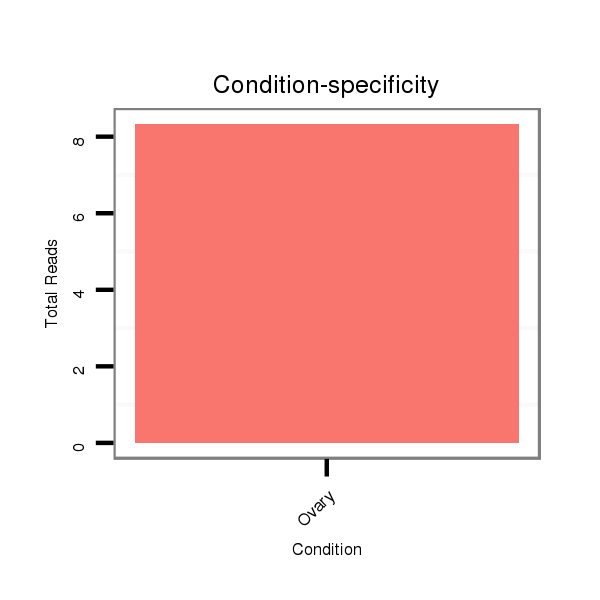
ID:dsi_21244 |
Coordinate:4:220102-220485 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
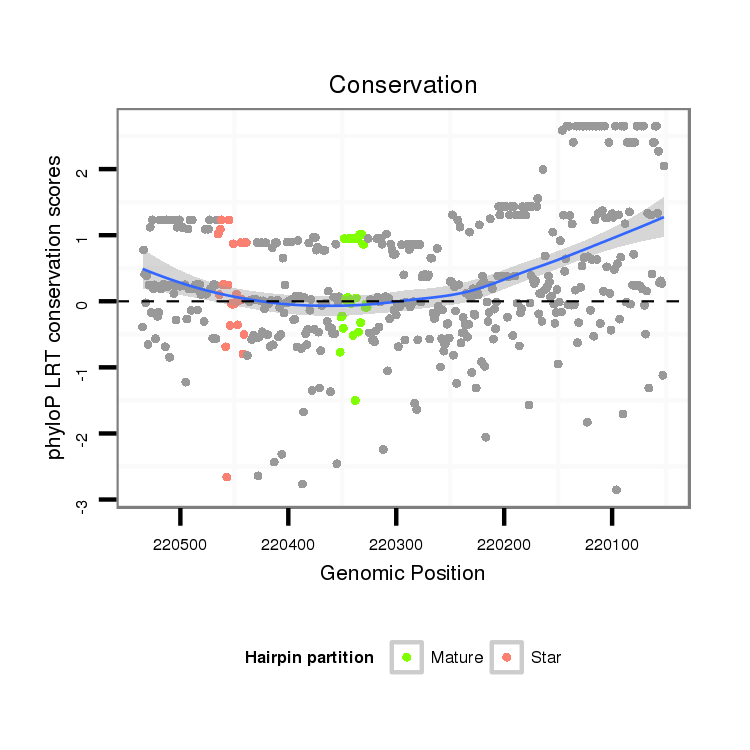
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.7 | -12.3 | -12.3 |
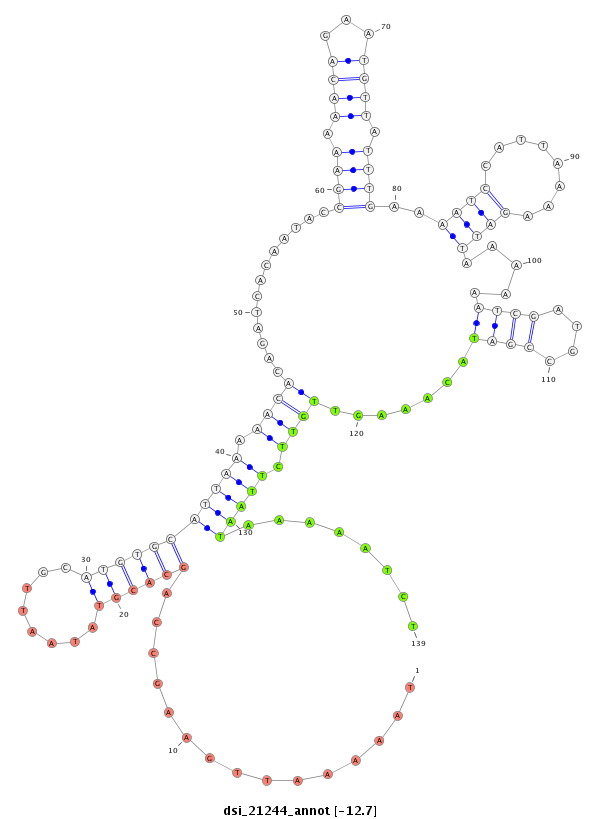 |
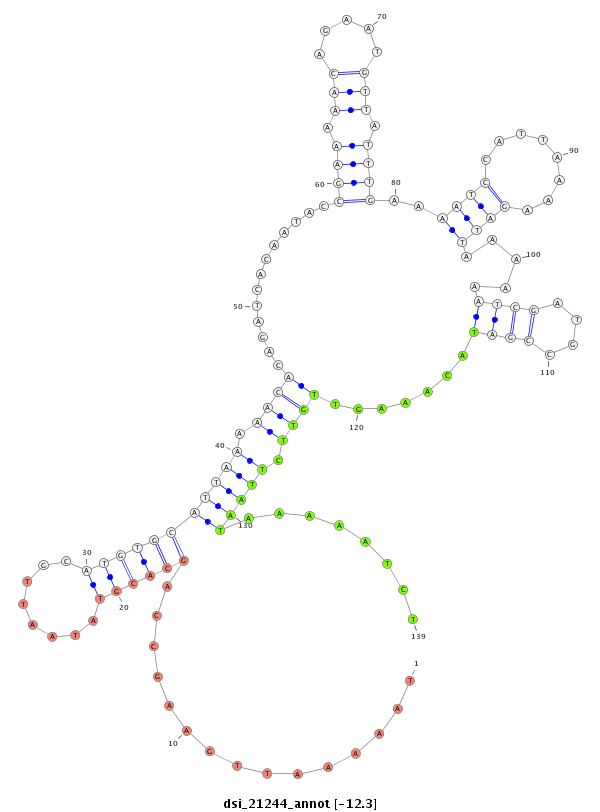 |
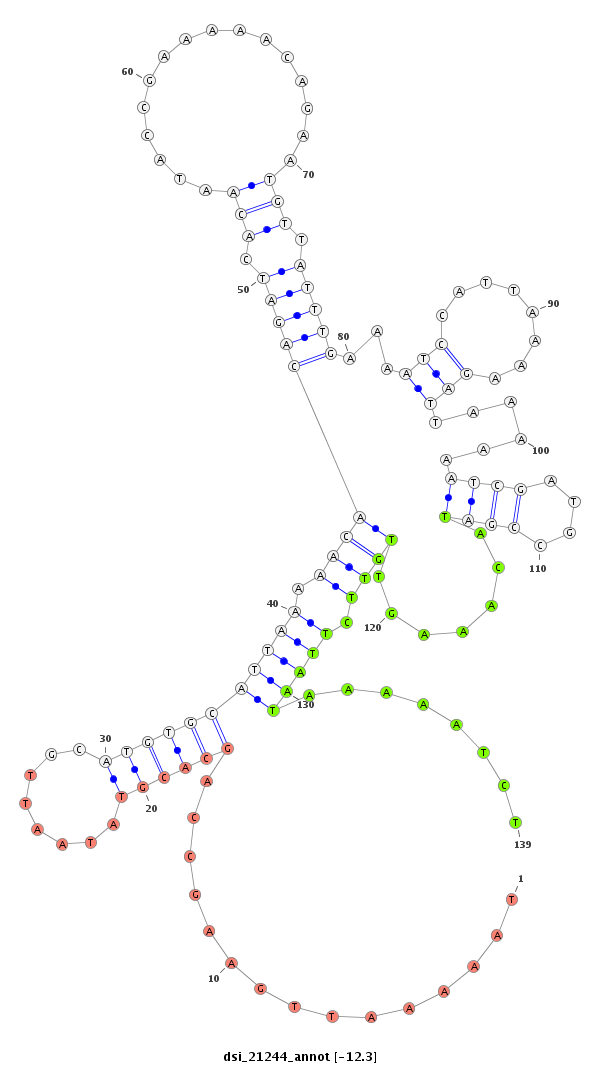 |
intergenic
No Repeatable elements found
|
TTGTTGAGCTTACCGTTGAAATAATAATTGTCGATGCCAACGAAATAAGTGTAAAGACTGAAAGAGTTTATAAAAATTGAAGCCAGCACGTATAATTGCATGTGCATTAAAAACACAGATCACAATACCGAAAAACAGAATGTTATTTGAAAATCCATTAAAAGATTAAAAAATCGATGCCGATACAAAGTTGTTCTTAATAAAAATCTTACGCCAATACGTGTTCTCGTGTGTGCTTAATATCTAACTTATATATATACTTAACATTTATTATAACTACATAGGTATATGGAATTTCACTCTAAGTAATGCATATCTTTGAAAGACAAATGAATTAAAGCAACATGTCGCGTGGCAAACATGCCACATTCTTAAATAATTCGCCAATTAAAATCAAACCATTCATTATATTAGTCTTGAATTTAAATATATGGTGTCAATATATTAGTGCAACTTCATATTTAAGCCTGAACGGTAAGATACA
**********************************************************************...............((((((........))))))(((((.(((((((((.(((................))).)))))..((((........)))).....((((....)))).......)))).)))))........*********************************************************************************************************************************************************************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....TGAGCTTACTGTTGAA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 16 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TACAAAGTTGTTCTTAATAAAAATCT................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................TAAAAATTGAAGCCAGCACGTATAATT................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................ATATTTAAGCCTGAACGG......... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................TGTTATTTGAAAATCCATTAAAAGA............................................................................................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................GTGTAAAGACTGAAAGAGTTT............................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TTAAAAACACAGATCACAATACCGAAAA.............................................................................................................................................................................................................................................................................................................................................................. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................TTAAAAACACAGATCACAATACCGAAAAA............................................................................................................................................................................................................................................................................................................................................................. | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................TTACGCCAATACGTGTTCTCGT.............................................................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AAAAATCGATGCCGATACAAAGTTGTTCT............................................................................................................................................................................................................................................................................................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................TCACTCTAAGTAATGCATATCTTTGA.................................................................................................................................................................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................TGAAAAGCAAATGAATTAAAGAAA............................................................................................................................................. | 24 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....TGAGCTTACTGTTGA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 15 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................GATTAAAAAATCGAT.................................................................................................................................................................................................................................................................................................................. | 15 | 0 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................AAGCTACATGTCGCG.................................................................................................................................... | 15 | 1 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTTCATATTCCAACCTGAAC........... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AATCGATGCCGATGCGAAGTC.................................................................................................................................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................TGGTTGTTCTTAATAAAAA...................................................................................................................................................................................................................................................................................... | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...TTGAGCTTACTGTCGA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...TTGAGCTTACTGTCGAA................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 17 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................CGTAGATGGAATCTCACT....................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................CTAAAAGAGTTTATGAAAA........................................................................................................................................................................................................................................................................................................................................................................................................................ | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................AGACTAAAAAGTCGAAGCC............................................................................................................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
|
AACAACTCGAATGGCAACTTTATTATTAACAGCTACGGTTGCTTTATTCACATTTCTGACTTTCTCAAATATTTTTAACTTCGGTCGTGCATATTAACGTACACGTAATTTTTGTGTCTAGTGTTATGGCTTTTTGTCTTACAATAAACTTTTAGGTAATTTTCTAATTTTTTAGCTACGGCTATGTTTCAACAAGAATTATTTTTAGAATGCGGTTATGCACAAGAGCACACACGAATTATAGATTGAATATATATATGAATTGTAAATAATATTGATGTATCCATATACCTTAAAGTGAGATTCATTACGTATAGAAACTTTCTGTTTACTTAATTTCGTTGTACAGCGCACCGTTTGTACGGTGTAAGAATTTATTAAGCGGTTAATTTTAGTTTGGTAAGTAATATAATCAGAACTTAAATTTATATACCACAGTTATATAATCACGTTGAAGTATAAATTCGGACTTGCCATTCTATGT
***********************************************************************************************************************************************************************************************************************************************************************************...............((((((........))))))(((((.(((((((((.(((................))).)))))..((((........)))).....((((....)))).......)))).)))))........********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M025 embryo |
SRR553486 Makindu_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................................................................................................................................GTATAGAAACTTTCTGTTTAC........................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................AGCTGCGGCTATGTTTTCACA.................................................................................................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................TCATGTTAGTATGGTAAGTAAT............................................................................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................AATTTTTTTGCTACGGCTAGT.......................................................................................................................................................................................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........AATGGCAACGTTATTACTAAA...................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................GTCCAGTGTTATGTCTTT............................................................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................ATTTTGTCTTTCAATAAAATTT............................................................................................................................................................................................................................................................................................................................................ | 22 | 3 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................................AGGTTGGTAAGTAAT............................................................................ | 15 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................TGTTTGTGTCTAGTGT........................................................................................................................................................................................................................................................................................................................................................................ | 16 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................ATAATTTTGTTGAAGTATAAAT.................... | 22 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................ACTTCGCTCGTGCAT........................................................................................................................................................................................................................................................................................................................................................................................................ | 15 | 1 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................AAATAATATTGATGTC.......................................................................................................................................................................................................... | 16 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................ATAGGGACGTTGAAGTATA....................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................TTTAGTTTCGTAAGTA.............................................................................. | 16 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 4:220052-220535 - | dsi_21244 | TTGTT--GAGCTTACCGTTGAAATAA-TAATTGTCGATGCCAACGAAATAAGTGTAAAGACTGAAAGAGTT-TATAAAAATTGAAGC-CAGCACGTATAATTGCATGTGC-----------ATTAAAAACACAGATCAC-AATACCG-----AA------AAACAGAA-------TGTTATTTGAAAATCCATTAAAAGAT-T---------------------------------------------------------------------------------------------------------------------------------------------AAAAAATCGATGCCGATACAAAGTTGTTCTTAAT------A----AAAATCTTACGCCAATACGTGTTCTC-G---------------------------------------------------------------------TGTGTGCTTAATATCTAACTTATATATATACTTAACATTTATTATAACTACATAGGTATATGGAATTTCACT--------------------------------------------------CTAAGTAATGCAT--------------------ATCTTTGAAAGACAAATGAATTAAAGC-AAC---ATGTCGCGTGGCAAACATGCCACATTCTTAAATAATTCGCCAAT---TAAAATCAAACCATTCATTATATTAGTCTTGAATTTAAATATATGGTGTCAATATATTAGTGCAACTTCATATTTAAGCCTGAACGGTAAGA-T-----ACA |
| droSec2 | scaffold_30:27500-27983 - | TTGTT--GAGTTTACCGTTGAAATAA-TAATTGTCGATGCCAACGAAATAAGTGTAAAGACTGAAAGAGTT-TATAAAAATTGAAGC-CAGCACGTATAATTGCATGTGC-----------ATTAAAAACACAGATCAC-AATACCG-----AA------AAACAGAA-------TATTATTTGAAAATCCATTAAAATAT-T---------------------------------------------------------------------------------------------------------------------------------------------AAAAAATCGATGCCGATACAAAGTTGTTCTTAAT------A----AAAATCTTACGCCAATACGTGTTCTC-G---------------------------------------------------------------------CGTGTGCTTAATATCTAACTTATATATATACTTAACATTTATTATAACTACATAGGTATATGGAATTTCACT--------------------------------------------------CTAAGTAATGCAT--------------------ATCTTTGAAAGACAAATGAATTAAAGC-AAC---ATGTCGCGTGGCAAACATGCCACATTCTTAAATAATTCGCCAAT---TAAAATCAAACCATTCCTTATATTAGTCTTGAATTTAAATATATGGTGTCAATATATTAGTGCAACTTCATATTTAAGCCTGAACGGTAAGA-T-----ACA | |
| dm3 | chr4:273440-273925 - | TTGTT-AGAGCTTACCGTTGAAATAA-TAATTGTCGATGCCAACGAAATAAGTGTAAAGACTAAAAAAAGTATATAAAAATTGAAGC-CAGCAAGTATAATTGTATGTAT-----------ATTAAAAACACAGATCTA-AATACCG-----AA------AAACAGAA-------TATTATTTGAAAGTCCATTAAAAGGA-T---------------------------------------------------------------------------------------------------------------------------------------------AAAAAATCGATGCCGATACAAAGTTGTTCTCAAT------A----AAAATCTTGCGCCAATACGTGCTCTC-G---------------------------------------------------------------------CGTGTGCTTAATATCTAACTTAATTATATACTTAACATTTATTATAACTACATAGGTATATGGAATTCTACT--------------------------------------------------CTAAATAATGCAT--------------------ATCTGTGAAAGACAAATGAATTAAAGC-AAC---ATGTCGCGTGGCAAACATGCCACATTCTTGAATAATTCGCCAAT---TAAAATCAAACCATTCATTATTTTAGTCTTGAATTTAAATATATGGTGTCAATACATTAGTGCAACTTCATATTTAAGCCTGAACGGTAAGA-T-----ACA | |
| droEre2 | scaffold_4512:1027585-1028090 + | TTGATTAGAGCTTACCATTGATATAA-TGATTGTCGATGCCAACAAAATACATGTAAGAACTGAAAGCTTTTTATAAAATTAAAAGC-CAGAAAATATAATTGTATGCAT-----------GTCAAAAACATAGATCTC-AAAACTTA----AAAACGTTAAACAGAA-------TATTATTTTAAAATCCAGTAA-AGGT-T---------------------------------------------------------------------------------------------------------------------------------------------AAAAAATCTATGCTGACATAAAGTTGTTCTAAATAAATATAAATAAAAATCTTGCGTC-ATAAGTGTTCTC-G---------------------------------------------------------------------CGTGTGTACAATATTTAACTTAATTCAATACTTAACAGTTAACATAATTACATAGGTATGTGGAGTTCTATT--------------------------------------------------CAAAGTGAAGCATATGT----------------ATCTGTGAAAGACAAATGAATTAAAGC-AAC---ATGTCTCGTGGCAAACATGCCACATTTTCGGATAATTCGCCAAT---TAAGATCAAACCAATCATTATTTTAGTCTTGAATTTAAATATTTGGTGTCAGAATATAAGTGCATCTTCATACTTAAGCTTGAATGGTAAGA-T-----ACA | |
| droYak3 | 4:278068-278560 - | TTGATAAGAGCTTACCATTGAAAAAA-TGATTGTCGATGCCAACGAAAGACGTGTTAGAAGTGAAAGCATTTTATAAAAATGCAAGC-CAGCTAAAACTATTGTATTTAT-----------ATGAAATACACCGATCTT-AAAACCA-----AAAACTTTAAACAGAG-------AA---TATATAAATCCATTAAAAGGT-T--------------------------------------------------------------------------GGAAT------------------------------------------------------------------TAGGGAGGCTGTTTCAAAGTTGTTATAAATAA----A----AAAATCTTGTGTCAATCAGTGTTCTT-G---------------------------------------------------------------------TGTGTGTATATTATTTAACTTAATTTAATACTTAACATTTAACATAATTATGTTGGTATGTGAAATTCTATT--------------------------------------------------CAAAGGAAAGCAT--------------------ATCTGTGAAAGACAAATGAATTAAAGC-AAC---ATGTCTCGTGGCAAAAATGCCACATTTTCGGATAATTCGCCAAT---TAAGATCAAACCATTCATTATTTTAGTCTTAAATTTAAGTATATGGTGTCAGAATATTAGTGCAGCTCCATATTTAACCCTAAATGGTAAGA-T-----ACA | |
| droEug1 | scf7180000408984:880779-881251 + | TTGAAA-AGGCTTA-TACTAAACCAA-ATTTTGTAGATACCAAG--CATAAGTGAAAGAGCTGAAAGCTTCTAA---AAAACAAA-----GCAAATATATTTTTATGTTC----------TATAGAATACATAGGCCGC-AAATGTA-----AT------A--------AAAACGCTCCATTCTCTTATCCAATAA--TAT-TAAATGTATAGTGCTGGAAATATTTGGAAAGTTAAACTTTACTTACTCACT-TAC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTACTTAGTGGCGCTAATATGTGAAATTCTTTTACTTTAATATTTTCTTAAATATCATTATAAGACTTTAAAAGTTTTTCTGTAAAAATAAGGCAT--------------------GTCGGTGAAAGACCTATGAATTAATAC-AAT---ATGACGGTTGGCAATAATGCCACATATTCGGATAATTTCCCAAT---TAGAATCAAACCGTTCGTTATTTTGATCTTGAATGTCAACATATGCTTTCATCATGTCAGCGCATCTTCTTTTTTAAACCTGAACGGTAAGA---------- | |
| droBia1 | scf7180000301495:17389-17874 - | ATGAAAAAAGCTTAATGCTGAAAAAA-T-ATTATCGATACCAACTACAGAAGTAGAGGAGTTGAAAGCTTTTTATAAAAATACAAGC-AATCAAATATTCTTTTCAGTACA-------TATATAAGAAGTACTTTTCGAAAAAGCCTTAAAC----CTTAA-------AAA-----------CCT---------------------------------------------------AACTTTATTTATAAGCT------------------------GAACT-----T----------------------AGCATTCATTGGAAGCCTATATATAGAGTGTT--AAAAAAAGTTTTCCGATATTAATTTCGTGTTAACTTATGTA----TAAATCTTTCGCGACTATGTGACATTTT--------------------------------------------------------------------TCG------------------------------------------TGGGAGAAAAGGTGTATTAAA-------------------------------------------------------------ATAATACAA---------TTTTAAAACTTATCGTT-----GTAAATGAATTAATAC-AAC---ATGCCGCGTGGCAACAATGCCACTTACTCGGATAATTCCTCAAT---CAAAATAAAACCATTCATTATTTTGATCTTAAATTTTAACATATGCCTTCAGCAAATTAATGCATCTTCATTTTTAAATTTCAACGGTAAGA-T-----ATA | |
| droTak1 | scf7180000413465:8806-9285 - | ATGAAACAAGCTTACTGCTGAAAAAATTTATTATCGATACCCAGTACATAAGTGAAAGAGCTGAAAGCTTTTAATGAAAATATGAGC-CAGCAAATATACTTATATTTACA-------TATATCTAAAATACAGGTCGTGAAAACCGCAAAT----CTATA-------AAA-----------AAC---------------------------------------------------AACTTTATCTATTCGCTGTATTATACTTGTGTGGAAGTGTTGAATTAAAT-------------------------------------------------------------CAAAGTGTTCGGATCTTAAGCTCGTCTTAATATA---------------TGCATATATATGTGCTATC-G--------------------------------------------------------------------TCGTGGGCACAATG-CTAACTTAAATACATA-------------------GAA----CATATG----------------------------------------------------------------------------TATTATGATAAAAAGTAATCGCT-----GTAAATGAATTAATAC-AAC---ATGACGCGTGGCAATATTGCCACACACTCGGTTAATTCCTCAAT---TAAAATCAAACCATTCATTATTTTGATCTTAAATTTTAACATATGCTTTCACCATATCAGTGCATCTTCGTTTTTAAACCTCAACGGTAAGA-C-----AAA | |
| droEle1 | scf7180000491238:1135474-1135900 + | TTGAAAAGAGTTCATTGCTGAAAAAA-TTATTGTCGATACCAAATACATAAGTGAAAGAGCTGAAAGCTTTTTA--AAAATAAAACTTCA--AC----T-----------GCTTATCTTAT---------------------------------------TCACGAAA-------TATCATTTGAAGGCCAATTTAAAGGGTC--------------------------------------------------------------------------GAAAC------------------------------------------------------------------TAAGTTTCCAAACATTAAGTTCTTCTCAACTT----A----TAAAGTGTGCGCGGGTATATGGT--------------------------------------------------------------------------CG---------TG-TCAACCTAAATGCATT-------------------GAT----TATGCGAAATATT-CT--TTTGATATTTTAT-AAAT---A---------------------GC---------------T--------------------GTTATTTTAAGGCAAATGAATTAATAC-AAT---ATGACTCGTGGCAACAATGCCACCTACTCGGATAGTTCCCTTGT---TAAAATCAAGCCATTTATTATTTTGATCTTGAATTTTAACATATGCTTTCACCATATAAGCGCATCTTCATTTTTAAACCTGAACGGTAAGA-T-----GCA | |
| droRho1 | scf7180000770171:10986-11408 + | TTGCAAAAAGCTCATTGTTGAAA-AA-TTATTGGCGATACCAAATACATAAGTGAAAGAGCTGAAAGCTTTTTATAAATATAAAAC------------T-----------ATTTATCTTAT---------------------------------------ACACAAAA-------TAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAATTTTCGAATATAACGTTCTTCTTTATTT----A----TAAAGCGTGCGCGGATATATGATATT-G--------------------------------------------------------------------TCGTGTGCACTATG-TTAACCTAAGTGCATA-------------------TAT----TATGCAGAATCTT-CT--CTTAATATTTTAT-AAATAATA---------------------GCTGCTAAAGTAGAGCAT--------------------TTTGTTGAAAGACAAATGAATTAATAC-AAT---ATGACGCGTGGCAATAATGCCACATACTCGGATAATTCCCTAGT---TAAAATCAAGCCATTCATTATTTTAATCTTGAATTTTAACATATGCTTTCATCATATAAGTGCATCTTCATTTTTAAATCTGAACGGTAAGA-T-----ATA | |
| droFic1 | scf7180000454086:62386-62799 - | TTGAACAGAGCTTTTTGCTGAAAAAC-TGATTGTGGATACCAGCTACATAAGCGAAA--------AGCTTTAAATAAAAATACAAT------------T------------TATATCTTAT---------------------------------------ACATAAAA-------TATAAATTGAAAGACACGTGAAAGTG--------------------------------------------------------------------------TTAAAACAAATAAAAAAAAAGAGAATATCACAATCAG---------G--------------------ATAAAAAG----------CAT---------------------A----AA---------------------ATT-GTTGTGCAGACTATCCATACAATCAAATTGTATTAATATCCCGGAAAAGGACGCTTATTAACTAAATCGTC----------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGCGTAAAGCCAATGAATTGAAAC-AAT---ATGACGGGTGGCAACAATGCCACAAACTCGGAAAACTCCCCGGT---TAAAATCAAGCCGTTTCTTTTTATAATCCTAAACTTTAATATATGTTTTAACCATATCAGTGCATCTTCATTTCTAAACCTAAACGGTAAGA---------- | |
| droKik1 | scf7180000302373:188102-188316 + | TATAAA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGGACAACTATTTGGAATACACCG--------------------------------------------------CTACATTTTGAAT--------------------AATAAACAATGATTGTTGAACTAAAACCAAAATGACGACCCTTGGCAAAAATAATACATATTCGACCCATTATTCTGTTAGCAAAATCAAGTCATTACTTTTTTTTATATTAAATTTCAACATATGCCTTCAGCATATCAGTGCTTCAACATATTTAAACCTGAACGGTAAG----------- | |
| droAna3 | scaffold_13045:548271-548363 - | TAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AATTAAACCACTAATACTTTTATTATTAAATTTAAACATATGCATTGAACAAATTAGTGCAACTTCATATCTTATTCCAAGTGGTAAGA-A-------- | |
| droBip1 | scf7180000395521:5103-5217 + | AA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TACCTCAATTTT---AAAAATAAAGCCATTAATACTTATTTTATTAAATTTAAACATATGCATTGAACAAGTCAGTGCAACTTCATTTCTTATTCCTCATGGTAAGA-TGTTAAACA | |
| dp5 | Unknown_group_62:3566-3684 - | A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGTGCTTGGATTTTTCCTTCAA---CAAAATTAGACCTATAATTTTGTTGATTTTGAATTTTAATATATGTGTTCAACATATAAGCGCAACTTCATTTTTGAACATAAATGGTAAGC-T-----ATA | |
| droPer2 | scaffold_42:92252-92370 + | A------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CGTGCTTGGATTTTTCCTTCAA---CAAAATTAAACCTATAATTTTGTTGATTTTGAATTTTAATATATGTGTTCAACATATAAGCGCAACTTCATTTTTGAACATAAATGGTAAGC-T-----ATA | |
| droWil2 | scf2_1100000004943:1177451-1177544 + | ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAGGTCAAACCAATCATATTTTTGATCTTAAATTTTAATATTTGTCTTCATCATATCAGTGCATCTTCATTCTTAAATATCAATGGTAGG--T-----GCA | |
| droVir3 | scaffold_13052:749010-749121 + | C-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCGGATAATT---TAAA---TAAGTTCAAGCCGATCATTTTTTTTATCCTGCACTTAAATAGTAGTATCCAGCAGATAAGCGCATCATCGTTCTTAATAATCAATGGTAAGGAT-----AAA | |
| droMoj3 | scaffold_6498:1456043-1456144 - | AA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAATTTGAA---TAAGTTCAAGCCGATTCTCTTTTTCATATTGCATTTAAATATTAGTATTCAGCGAATCAGCGCATCGTCATTTCTCATTATCAATGGTAA------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/18/2015 at 12:17 PM