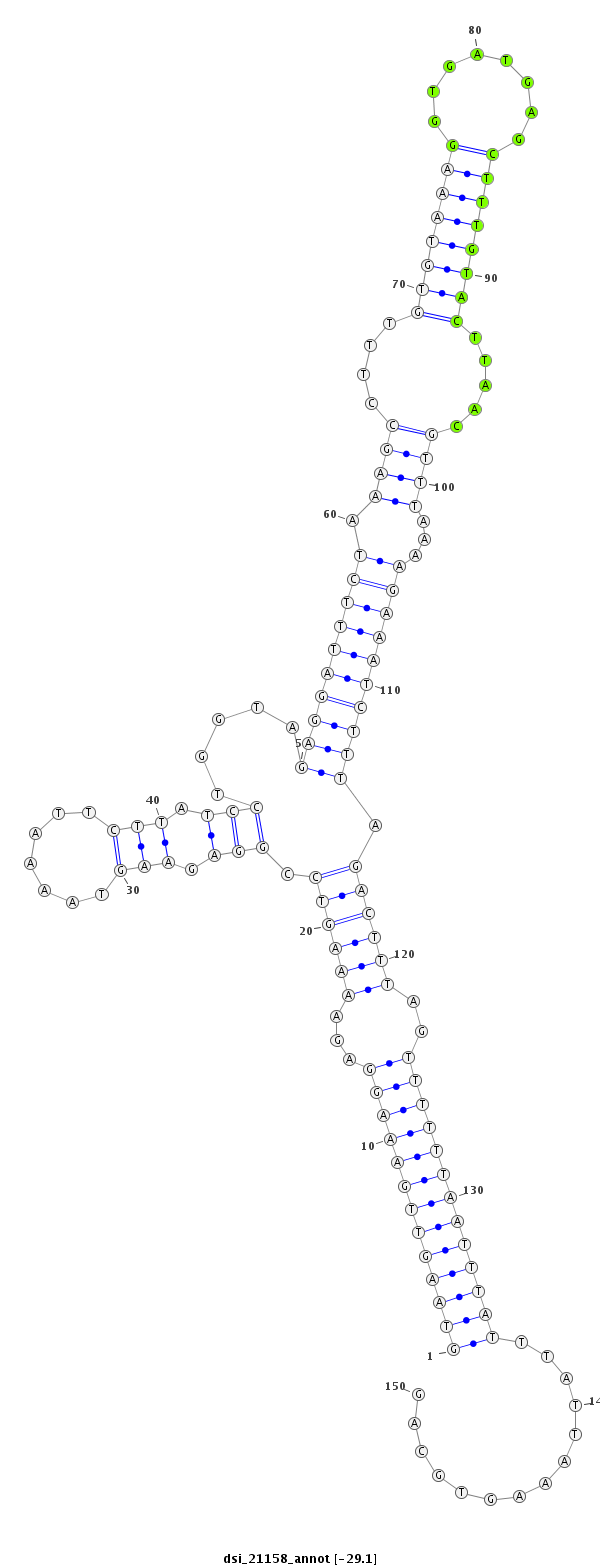
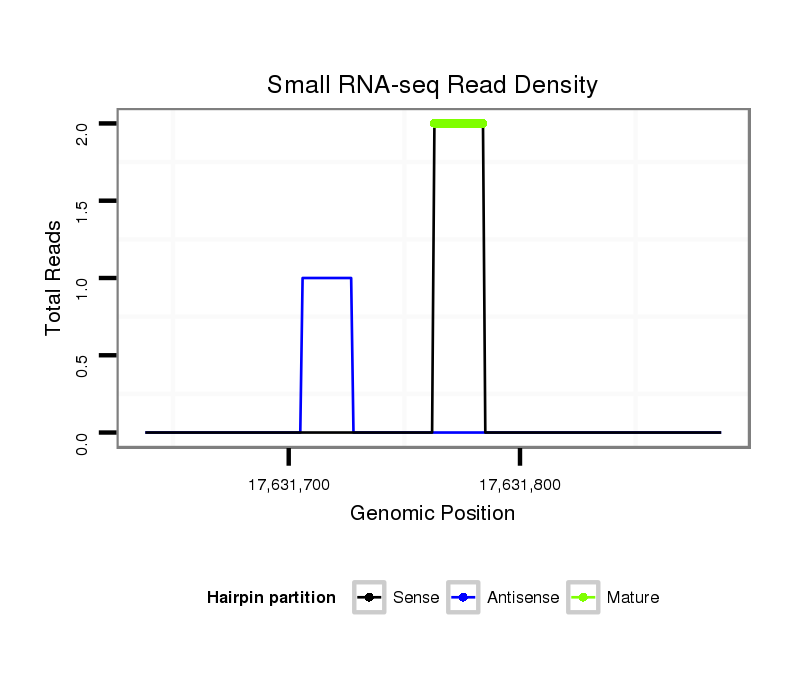
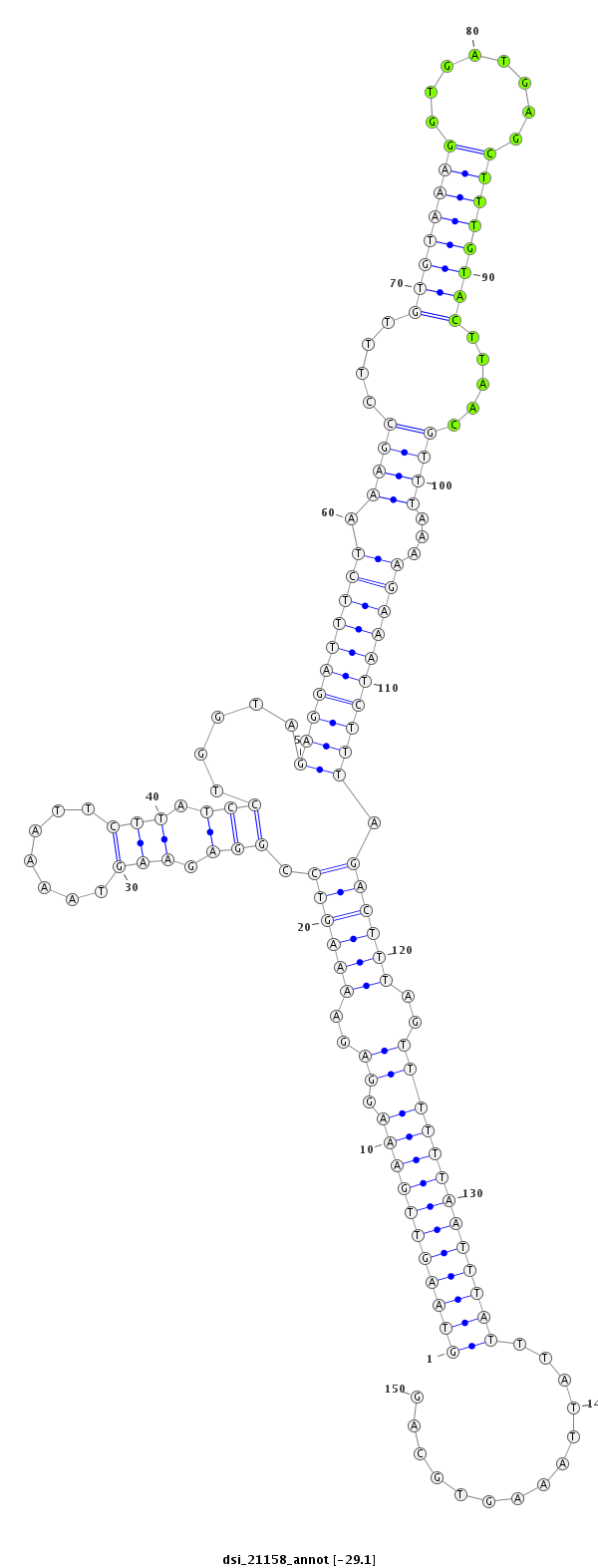
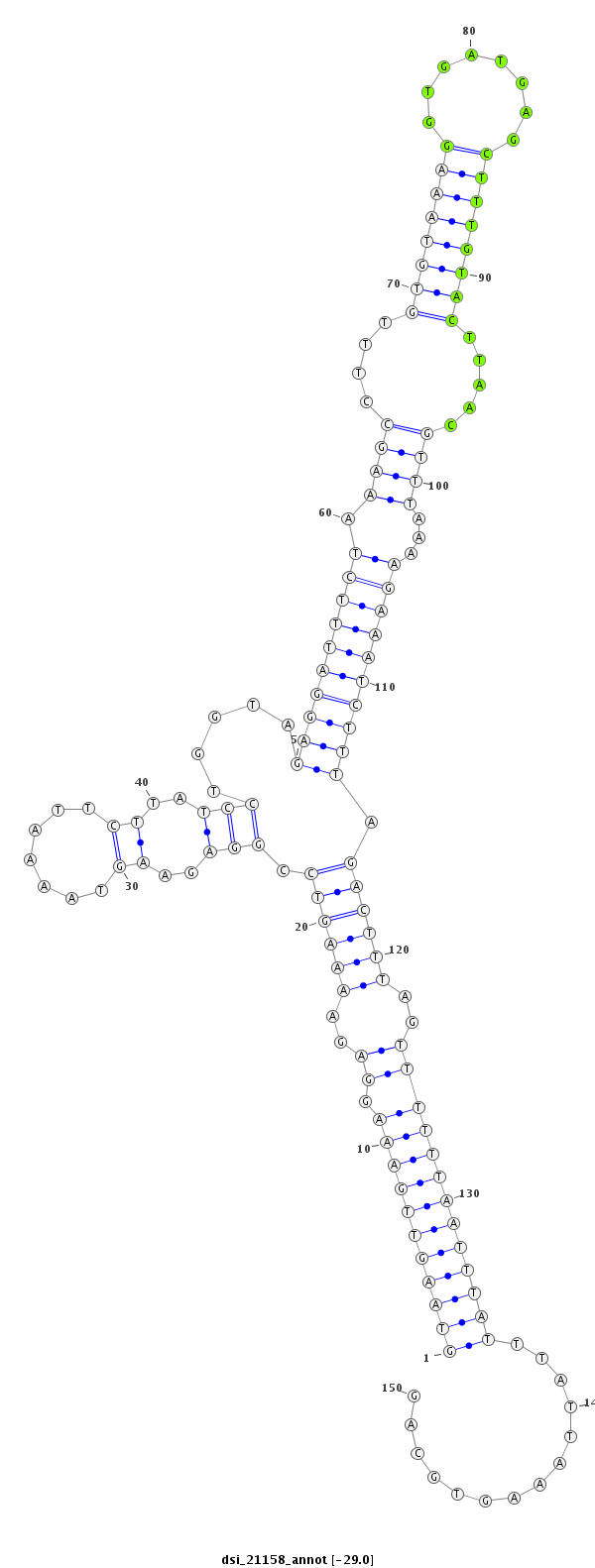
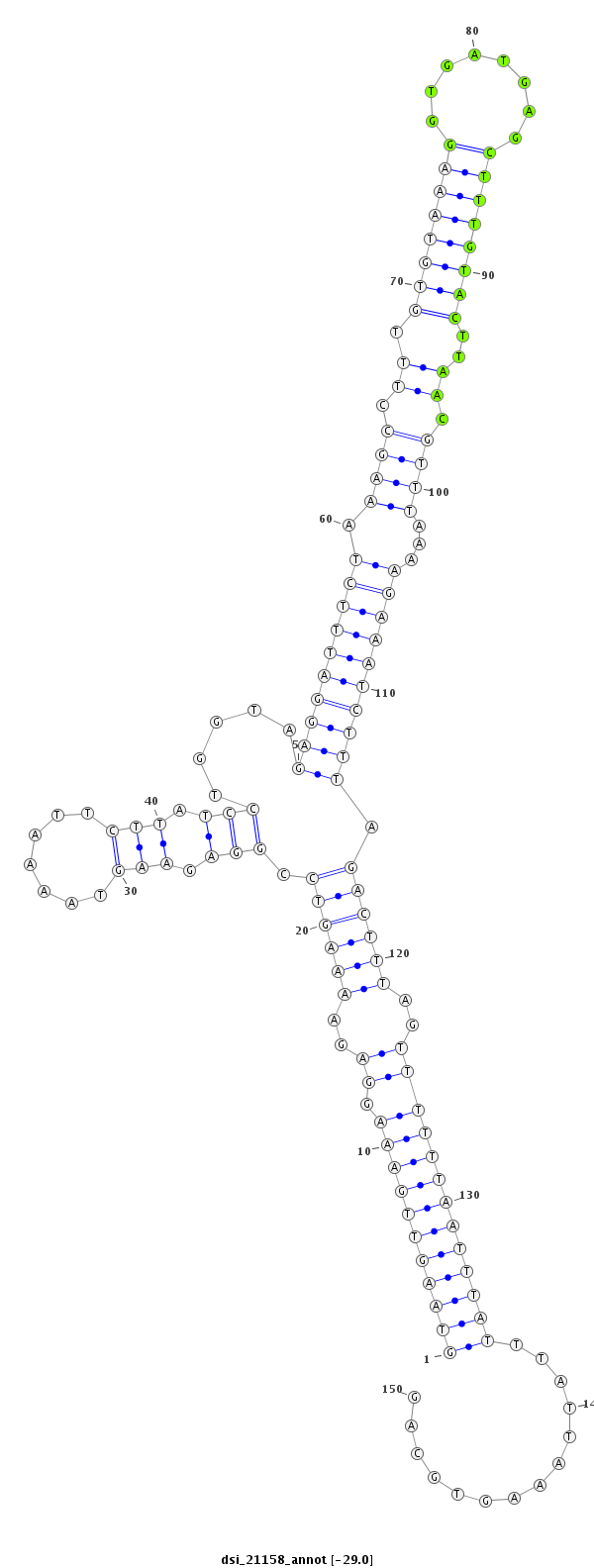
ID:dsi_21158 |
Coordinate:3r:17631688-17631837 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -29.1 | -29.0 | -29.0 |
 |
 |
 |
intergenic
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
|
CTGCCTGAGTCCGCGCTCAGCTTATTTCTACGAATTTCCACCGACAGCTGGTAAGTTGAAAGGAGAAAAGTCCGGAGAAGTAAAATTCTTATCCTGGTAGAGGATTTCTAAAGCCTTTGTGTAAAGGTGATGAGCTTTGTACTTAACGTTTAAAAGAAATCTTTAGACTTTAGTTTTTTAATTTATTTATTAAAGTGCAGCTGCGAAGCTGCAGTAAATGTAGAGCACAATTGTAATGAGGTCAGAATTT
**************************************************(((((((((((((...((((((.(((.(((.......))).))).....((((((((((.((((....((((((((........)))))))).....))))...)))))))))).))))))..)))))))))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
SRR553488 RT_0-2 hours eggs |
SRR902008 ovaries |
SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................GGTGATGAGCTTTGTACTTAAC....................................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................TCTTAGCCTGGTAGA..................................................................................................................................................... | 15 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................TGCAGAAAATGTAGAGCACAAG................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................TTGTAATGAGGTCAGAGCT. | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................CTTAGCCTGGTAGAG.................................................................................................................................................... | 15 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................TAGGATTTCTAGAGCCT...................................................................................................................................... | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................TGTGGAACGGTGATGAGCC.................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................TAGTTGTAATGAGGTCGGA.... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................AACGTCCGGAGACGTAA....................................................................................................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................CTTAGCCTGGTAGAGC................................................................................................................................................... | 16 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................AGCCTGGTAGAGTATT................................................................................................................................................ | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................AAAGAAATCTTCAGA................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................AAATCTTTAGCCTTT............................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................GCTGGAAGGTTAAAAGGAG......................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GACGGACTCAGGCGCGAGTCGAATAAAGATGCTTAAAGGTGGCTGTCGACCATTCAACTTTCCTCTTTTCAGGCCTCTTCATTTTAAGAATAGGACCATCTCCTAAAGATTTCGGAAACACATTTCCACTACTCGAAACATGAATTGCAAATTTTCTTTAGAAATCTGAAATCAAAAAATTAAATAAATAATTTCACGTCGACGCTTCGACGTCATTTACATCTCGTGTTAACATTACTCCAGTCTTAAA
**************************************************(((((((((((((...((((((.(((.(((.......))).))).....((((((((((.((((....((((((((........)))))))).....))))...)))))))))).))))))..)))))))))))))..............************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
M025 embryo |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
M023 head |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................ACGCTTCGACGTCATTTCCAA............................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................TCAGGCCTCTTCATTTTAAGAA................................................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACGTTTCGATGTCATTTCCA............................. | 20 | 3 | 8 | 0.25 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................TCGACGTCATTTCCAT............................ | 16 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................ACGCTTCAACGTCATTTC............................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................ACTTCGACGTCATTTCCA............................. | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................AAGATTTCGGAAGCA.................................................................................................................................. | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................CGTTTCGATGTCATTTCCA............................. | 19 | 3 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................ACTTCGACGTCATTTCCAC............................ | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................................................................................................TCGACGTCATTTCCA............................. | 15 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................CGAATACAGCTGCTTA....................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
Generated: 04/23/2015 at 11:57 PM