
ID:dsi_21141 |
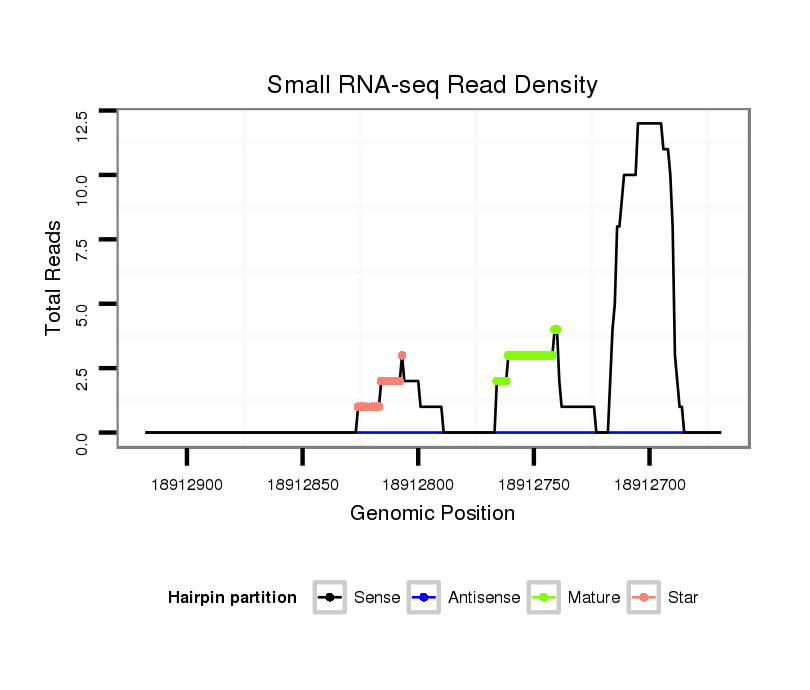
Coordinate:3l:18912719-18912868 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
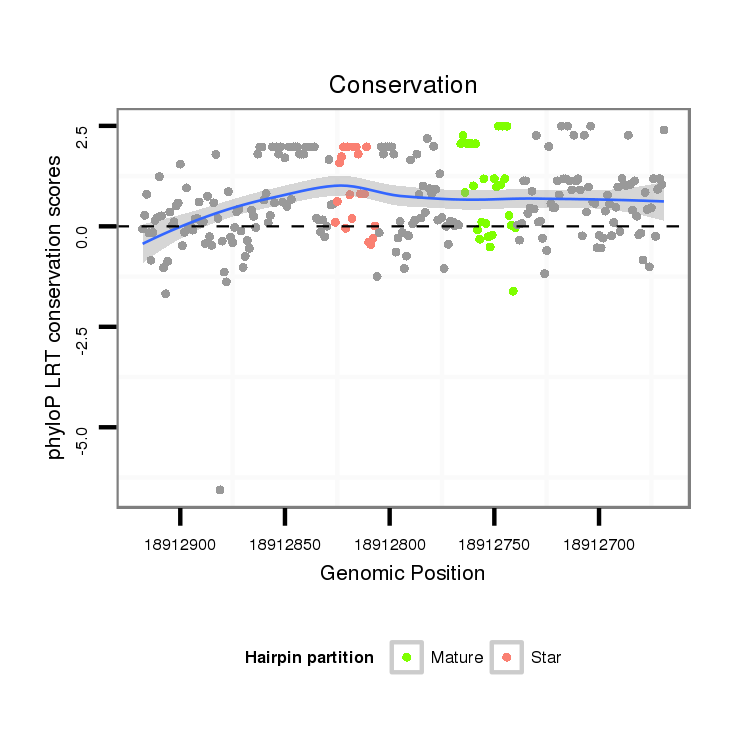
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -18.5 | -17.8 | -17.6 |
 |
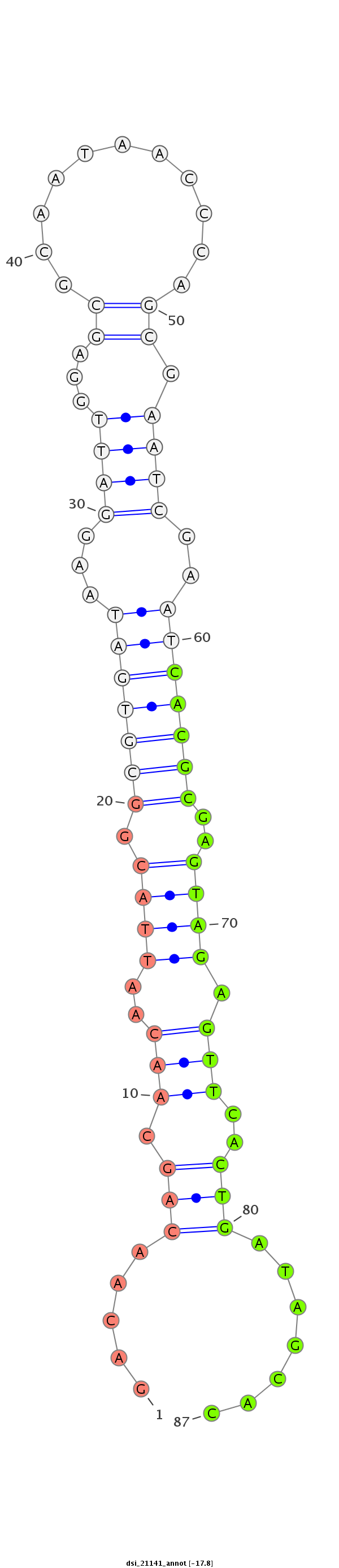 |
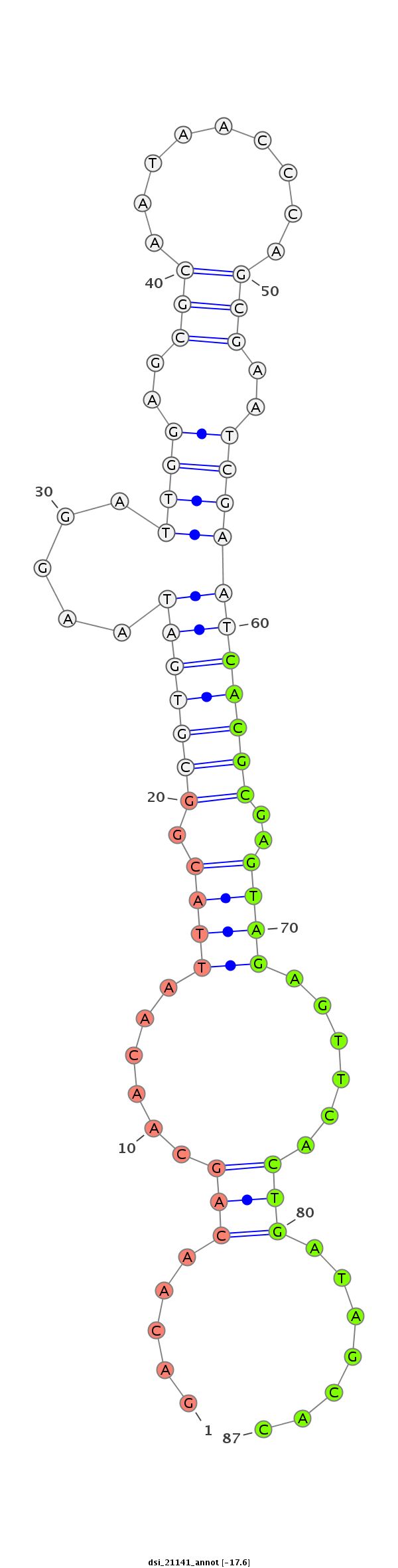 |
five_prime_UTR [3l_18912712_18912718_-]; CDS [3l_18912603_18912711_-]; exon [3l_18912603_18912718_-]; intron [3l_18912719_18913035_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCTTCCTCTTGTCTTAATCAGCCTCCCCCCCTGCCCCGCGACGCTCTCCTTTGATGATCAAAAAACAATCAGTTTGCGTAATATGCGCCAATGACAACAGCAACAATTACGGCGTGATAAGGATTGGAGCGCAATAACCCAGCGAATCGAATCACGCGAGTAGAGTTCACTGATAGCACTGCAACTCTTTTCCCTTCCAGATTTAGCATGACGGAATTCGAGGAGGACTTGCCCCACAACGGCCTGATGG ********************************************************************************************.....(((.(((..((((.(((((((.....((((..(((.........)))..)))))))))))..)))).)))..))).......*********************************************************************** |
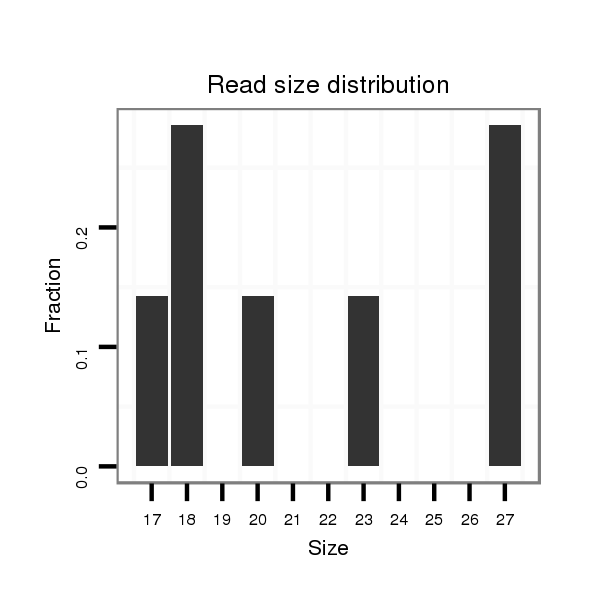
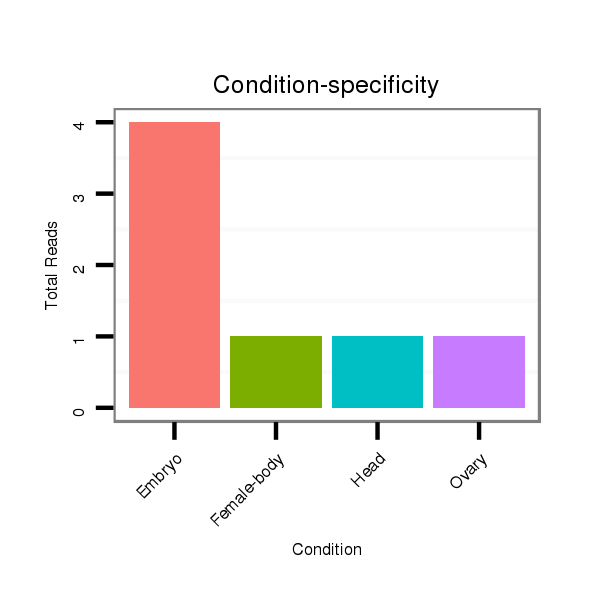
Read size | # Mismatch | Hit Count | Total Norm | Total | M025 embryo |
M024 male body |
M023 head |
SRR902008 ovaries |
M053 female body |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................CGTGATAAGGATTGGCTC........................................................................................................................ | 18 | 2 | 2 | 7.00 | 14 | 3 | 7 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AGCATGACGGAATTCGAGGAGGACT..................... | 25 | 0 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CACGCGAGTAGAGTTCACTGATAGCAC....................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TTTAGCATGACGGAATTCGAGGAGGAC...................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................ATGACGGAATTCGAGGAGGACTT.................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................GACAACAGCAACAATTACGG.......................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................TCGACGCTCTCCTTGGATG.................................................................................................................................................................................................. | 19 | 2 | 3 | 1.00 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| .........................................................................................................................................................................................................TTTAGCATGACGGAATTCGAGGAGGACT..................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GAATTCGAGGAGGACTTGCC................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................ACTGCAACTCTTTTCCCT....................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GAATTCGAGGAGGACTTG................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CATGACGGAATTCGAGGAGGA....................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TTAGCATGACGGAATTCGAGGA.......................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AGCATGACGGAATTCGAGGAGGAC...................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................ACAATTACGGCGTGATA................................................................................................................................... | 17 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................TAGCATGACGGAATTCGAGGAGGACT..................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................TGGCAAATTTAGCATGACGGAATTCGAGG........................... | 29 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TTAGCATGACGGAATTCGAGGAGGACT..................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GAGTAGAGTTCACTGATAGCACT...................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GCGGAATTCGAGGAGGAC...................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................GCGTGATAAGGATTGGAG......................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CTTCGTGATAAGGATTGG........................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................CTTTGATGATGAAAAGAC........................................................................................................................................................................................ | 18 | 2 | 7 | 0.29 | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................CGGGATAAGGATTGGCGC........................................................................................................................ | 18 | 2 | 9 | 0.22 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................CCGCCCCGCAAGGCTCTCCT........................................................................................................................................................................................................ | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................CGTCGGGATAAGGATTGG........................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................CCTTCCAGATTTAGG........................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................CTTCGTGATAAGGATTGGCG......................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AGCACTGCAACTCTTCCCCA........................................................ | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................CCCCCCGGCCCCGCG.................................................................................................................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...............................GGCCCCGCGCCGCACTCCT........................................................................................................................................................................................................ | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AGAAGGAGAACAGAATTAGTCGGAGGGGGGGACGGGGCGCTGCGAGAGGAAACTACTAGTTTTTTGTTAGTCAAACGCATTATACGCGGTTACTGTTGTCGTTGTTAATGCCGCACTATTCCTAACCTCGCGTTATTGGGTCGCTTAGCTTAGTGCGCTCATCTCAAGTGACTATCGTGACGTTGAGAAAAGGGAAGGTCTAAATCGTACTGCCTTAAGCTCCTCCTGAACGGGGTGTTGCCGGACTACC
***********************************************************************.....(((.(((..((((.(((((((.....((((..(((.........)))..)))))))))))..)))).)))..))).......******************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
M023 head |
SRR553485 Chicharo_3 day-old ovaries |
O001 Testis |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
M053 female body |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GTTAAGGCCCCACTATTC................................................................................................................................. | 18 | 2 | 4 | 4.00 | 16 | 8 | 5 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................TTTTTTTTTAGTCAAGCGCAT.......................................................................................................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................GTTAAGGCCCCACTATTCG................................................................................................................................ | 19 | 3 | 18 | 1.00 | 18 | 3 | 0 | 0 | 4 | 4 | 1 | 4 | 0 | 2 | 0 |
| .......................................................................................................GTTAAGGCCCCACTATT.................................................................................................................................. | 17 | 2 | 13 | 0.38 | 5 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 1 |
| .....................................................................................................TGGTTAAGGCCCCACTATTC................................................................................................................................. | 20 | 3 | 7 | 0.29 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................TCATGCCGGACTATTCTTAA............................................................................................................................. | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................TTAAGGCCCCACTATTC................................................................................................................................. | 17 | 2 | 13 | 0.15 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................GTTAAGGCCCCACTAT................................................................................................................................... | 16 | 2 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TGGTTAAGGCCCCACTATT.................................................................................................................................. | 19 | 3 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 0 |
| ......................................GCTGCGAGAGGGATCTA................................................................................................................................................................................................... | 17 | 2 | 8 | 0.13 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................CGGTGTGCTGCGAGAG.......................................................................................................................................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................TAAGGCCCCACTATTC................................................................................................................................. | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................AGGGAAGGTCGAAAT............................................. | 15 | 1 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................CAGGGCGCTGCGCGAGG......................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................GCTGCGAGAGGGAACTC................................................................................................................................................................................................... | 17 | 2 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................AGGGGGGGAAGGGGC.................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................AGGGAAGGTGGAAATCGTG......................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................TTAAGGCCCCACTATT.................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................GGAAACTACGAGTTTCTTA........................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................GAGCTGCGAGAGGGAA...................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3l:18912669-18912918 - | dsi_21141 | TCTTCCTCTT-GTCTTAATCAGCCTCCCCCCCTG-----CCCCG-CG----ACGCTCT--CCTTTGATGATCAAAAAACAATCAGTTTGCGTAATA-TGCGCCAATGACA---ACAG---CAACAA-TTACGGCGTGATAAGGAT-TGGAGCGCAATAACCCAGCGAAT-CGAATCA-CGCGAGTAGAG--------------------TTC-------------ACTGATA-GCAC----------TGCAACTCTTTTCCCTTCCAGATT-------TAGCATGACGG-------------AATTCGAGGAGGACTTGCCCCA--C-AACGGCCTGATGG |
| droSec2 | scaffold_19:1207585-1207833 - | TCTTCCTCTT-GCCTTAATCAGCCTCC-CCCCTG-----CCCCT-CG----GCGCTCT--CCTTTGATGATCAAAAAACAATCAGTTTGCGTAATA-TGCGCCAATGACA---ACAG---CAACAA-TTACGGCGTGATAAGGAC-TGGAGCGCAATAACCCAGCGAAT-CGAATCA-CGCGAGTAGAG--------------------TTC-------------ACTGATA-GCAC----------TACAACTCTTCTCCCTTCCAGATT-------TAGCATGACGG-------------AATTCGAGGAGGACTTGCCCCA--C-AACGGCCTGATGG | |
| dm3 | chr3L:19253281-19253530 - | TCTTCCTCTT-TTCTTAATCAGCCTCC-CCCTTG-----CCCCT-TG----ACGCACT--CCTTTGATGATCAAAAAACAATCAGTTTGCGTAATA-TGCGCCAATGACA---ACAG---CAACAA-TTACGGCGTGATAAGGAC-TGGAGCGCAATAACCCAGCGAATCCGAATCA-CGCGAGTAGAG--------------------TTC-------------ACTGATA-GCAC----------TGCAACTCTTTTCTCTTCTAGATT-------TAGCATGGCGG-------------AATTCGAGGAGGACTTGCCCCA--C-AATGGCCTGATGG | |
| droEre2 | scaffold_4784:19065409-19065651 - | TCTTCCTCTTTTTCTTAATCAGCCTCC-CC-CTG-----CCCCG-CAAC-----------CTAATGATGATCGACAAACATTAAGTTTGCGTAATA-TGCGAGAATAACA---ACAG---CAACAA-TTGTGGCCTGATAAGGAC-TGGAGCCCAATAACCCAGCGGAT-CGAATCA-CGCGAGTAGAG--------------------AT--------------ACTGATA-GCAC----------TGCAACTCTTTTCCGTTCCAGATT-------TAGCATGGCGG-------------AGTTCGCGGAGGTCTTGCCCCA--C-AGTGGTCTGATGG | |
| droYak3 | v2_chr3L_random_050:20580-20823 - | TCTTCCTCTT-TTCTTTATCAGCCTCCCCCCCTA-----CCCAG-CGC----------G-CTTTTGATGATCACAAAACAATCAGTTTGCGTAATA-TGCGACAATAACA---ACAG---CAACAA-TTACGGCGTGATAAGGAC-TGGAGCACAATAACCCAGCGAAT-CGAATCA-CGCGAGCAGAG--------------------TT--------------ACTGATA-GCAC----------TGCAACCCTTCTCCATTCCAGATT-------TAGCATGGCGG-------------AATTCGCGGAGGTCTTGCCCCA--C-AATGGCCTGATGG | |
| droEug1 | scf7180000409471:212983-213220 + | TCTTTCTCTT-CGCTTAATCT----TCCCCCTTC-CCT-CCCCC-CAAC-------CTC-AGTTGGATGATCAAAAAACAATCAGTTTGCGTAATA-TGCGACAATAACA---ATAA---CAACGT-ATTCGGGGTGATAAGAAC-TCGGGCACAATGAACCA-----G-CGA-ACA-CGCGAGTAAAG--------------------TT--------------ACTGATAAGCGC----------TGCAACTCTTTT---TTCCAGATT-------TAGCATGACGG-------------AATATACGGAGGTCTTGCCCCA--C-AATGGTATAATGG | |
| droBia1 | scf7180000302529:221505-221726 + | TC--------------AACCC----CC--G-CTCCCCTCCCCCGCCATC-----------CCCTCGACGATCAAAAGACAATCAGTTTGCGTAATA-TGCAACAATTACA---ACAACAATAACAA-TTGC-GCGTGATAAGGAC-TGAGGCACAATGAACCA-----G-CGGACCA-GGCGAGCA-AG--------------------TT--------------ACTGATAAGCCC----------TGCAACTCTTT-----TCCAGATT-------TAGCATGACGG-------------AACACGCGGAGGTCGTGGCCCA--C-AATGGCCTGATGG | |
| droTak1 | scf7180000414404:32085-32297 + | T--------------TAATCT----CT-ACCCTCCCC--G-CCACCATC-------CTC-CCTTTGATGATCAAAAAACAATCAGTTTGCGTAATA-----------ACA---ATAA---CAACAA-TTA-GGGGTGATAAGGGC-TCTGGCACAATGAACCA-----G-CGAGACA-CGCGAGTAAAG--------------------TT--------------ACTGATAAG--C----------TGCAACTCTTTT--TTCCCAGATT-------TAGCATGACGG-------------AATACACGGAAGTCTTGCCCCA--C-AATGGCCTAATGG | |
| droEle1 | scf7180000491249:6548468-6548685 - | TCTTCTTCTT-TGATTAATCT----TCCCCCCTC-----CCCCG-CACC-----------CGTGTGATGATAAAAAAACAATCAGTTTGCGTAATA-TGAGGCAACAGCA---ATAA---CAACAA-TTACTGGGTGATAAGGA---------CAATG--------------AACCA-CGCGAGTAAAG--------------------TC--------------ACTGATAATCGC----------TGCAACTCATTT--TTTCCAGATT-------TAGCATGACGG-------------AGTTCACGGAAATCTTGCCACA--C-AAGGGCCTAATGG | |
| droRho1 | scf7180000767296:135-363 - | TCTTCTTCTT-TGATTAATCT----TCCCCCCTCCTC--TCCTC-TGAC-----------CCCATGATGATAAAAAAACAATCAGTTTGCGTAATA-TGGGACAATAACA---ATAA---CAACAA-TTACTGAGTGATAAGGA---------CATTGAACCA-----T-TTAAACA-CGCGAGTAAAG--------------------TT--------------ACTGATAAGCAC----------TGCAACTCTTTT---TTTCAGATT-------TAGCATGACGG-------------AATTTACGGAAATCTTGCCCCA--CCAAGGGCCTAATGG | |
| droFic1 | scf7180000453824:641669-641905 - | CTTCTT-C---GCTTCAATCT----TCTCC-CTCCACT-CCCCT-CACC-----------ATTTCGACGATCAAAAAACAATCAGTTTGCGTAATA-CAAGACAATAACA---ATAA---CAACAA-TTATGGGATGATAAGGAC-TCAAGCACAATGAACCACCA--G-CGAGACA-CGCGAGTGAAG--------------------TGA-------------ACTGATAAGC-------------GCAAACCTTCTTTTTTCCAGATT-------TAGCATGACGG-------------AATTCGCGGAGGTCGTGCCCTA--C-TATGGATTAATGG | |
| droKik1 | scf7180000302686:883380-883628 - | TCTTCT---T-TTATTAATCCCCCACC-CCATT------CCCCT-TCCCCTTCCGTTTCGTTTGTGTTGATCAAAAAACAATCAGTTTGCGTAATAATGCGCCAATAACA---ATAA---CAACAA-TTAGCAAGTGATAAGCCC-TGCAGAACGTTGAACCA-----T-CGAAACA-CGCGAGTCAAG--------------------TG--------------ACTGATAAGCCT----------TGTAACTCTTT-CATTTCCAGATT-------TAGCATGACGG-AGG---------AATACTCGGAAATCTTGCCCCA--G-AGGGGTCTAATGG | |
| droAna3 | scaffold_13337:18204276-18204497 + | GAT-----------------AGCATCC-CC-CTG-----CTCCC-CT----TCCCTCTT-CTATCAATGATCAGAA-ACA---AATTTGCGTAATA-AGCGAAATTAACATAAATAATAACAACAAATT--GCACTGATAAGAGCCACGAGAACATTGAACCA-----C------CA-CGCGAGCCGAA--------------------CC--------------ACTGATAAGATG----------CCCAACTCTTTA---TTCCAGATT-------TAGCATGCCGG-------------AATACATGGAGGTCTTGCCCCA--C-CGCTCCCTGATGG | |
| droBip1 | scf7180000396741:1386775-1386995 + | TCTGCT-------------------CC-CC-CTC-----TCCCGCCCCC-------CTC-CTATCAATGATCAGAA-ACA---AATTTGCGTAATA-AGCGACATTAACATAAATAATAACAACAA-TTGC--ACTGATAAGGGCCCCGAGAACATTGAACCG-----T-CGG-ACA-CGCGAGCCGAA--------------------TC--------------ACTGATAAGATG----------TGGAACTCTTTA---TTCCAGATT-------TAGCATGCCGG-------------AATACATGGAGGTCTTGCCCCA--C-CGCGGCCTGATGG | |
| dp5 | XR_group8:7344929-7345171 - | TCTCGCTCTC-GCCCT---------CC-CA-CTC-----TCCCGCTCTC-------CCG-CTCTCAGCGATCAGGAAACAATCAGTTTGCGTAATA-TACA------------ACAA---TAACAA-TTA-AATGTGATAAGCCT-GGGGGCACATTGAACCA-----T-TAA-ACACCGCGAGTGAAT--------------------CCCAGCCATAACTATAACTGATAAGAGCCCTTACGTTTGTTAACTCTTTG----CCCAGATT-------TAGCATGACTG-------------AAAGTTTGGAGGACACACCCGA--T-GCGGGGCTGATGG | |
| droPer2 | scaffold_48:135094-135339 - | TCTCTGCATT-GCATCGCTCT----CGCCC-TCC--------CACTCTC-------CCG-CTCTCAGCGATCAGGAAACAATCAGTTTGCGTAATA-TACA------------ACAA---TAACAA-TTA-AATGTGATAAGCCT-GGGGGCACATTGAACCA-----T-TAA-ACACCGCGAGTGAAT--------------------CCCAGCCATAACTATAACTGATAAGAGCCCTTACGTTTGTTAACTCTTTG----CCCAGATT-------TAGCATGACTG-------------AAAGTTTGGAGGACTCACCCGA--T-GCGGGGCTGATGG | |
| droWil2 | scf2_1100000004540:544923-545053 - | A------------------------------------------------------------------------------------------------------------------------------------------------------------------GCGGCT---CAACA-CGCGGGTCAAAA-------------------CT--------------GATAACAAATAA----------CACAATTTCTTTAATTTTTACATTTTTTTAATAGCATGGCAT-CCGTGGATACGGAATTCAGAAGCAACTTGCCCCCATCCTCTGGTCTATTGG | |
| droVir3 | scaffold_13049:12836243-12836252 - | G-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACCCGTCAG | |
| droGri2 | scaffold_15110:23483343-23483458 - | GCGCGT------------------------------------------------------------------------------------------------------------------------------------------------------CAAATCAGCGCA----------------CATTGATTCACAGCAAATAGCACATGT--------------ACTGATA-ACGC----------TTCCC-----------TCCCCACC-------TCCCCCCGCAGAATATTTGCACGAAATTGAACATGGAACTGCCCCA----------------G |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 04:58 PM