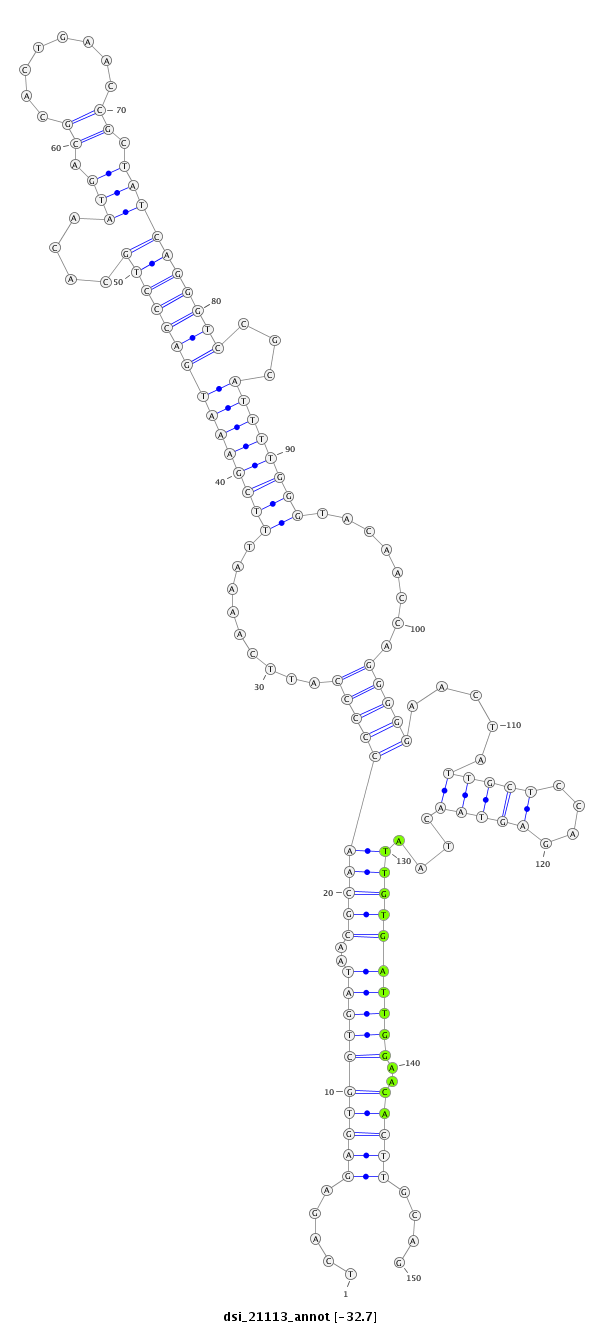
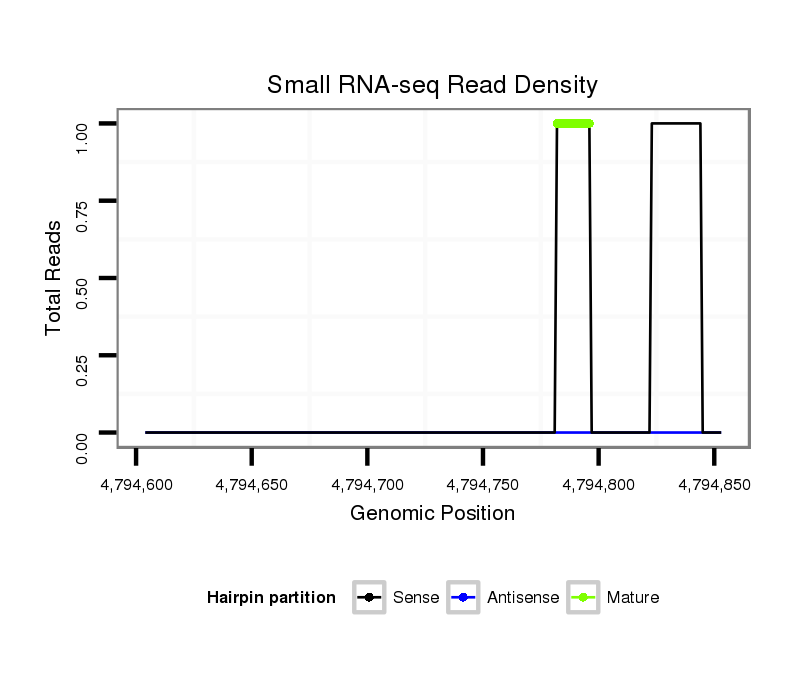
ID:dsi_21113 |
Coordinate:2r:4794654-4794803 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
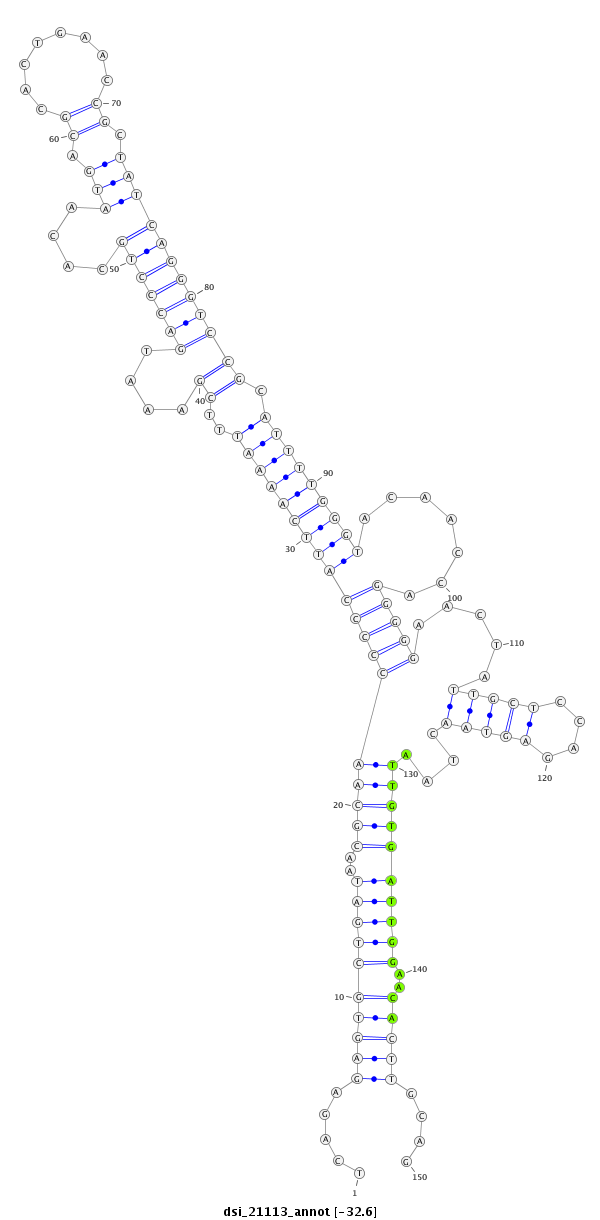
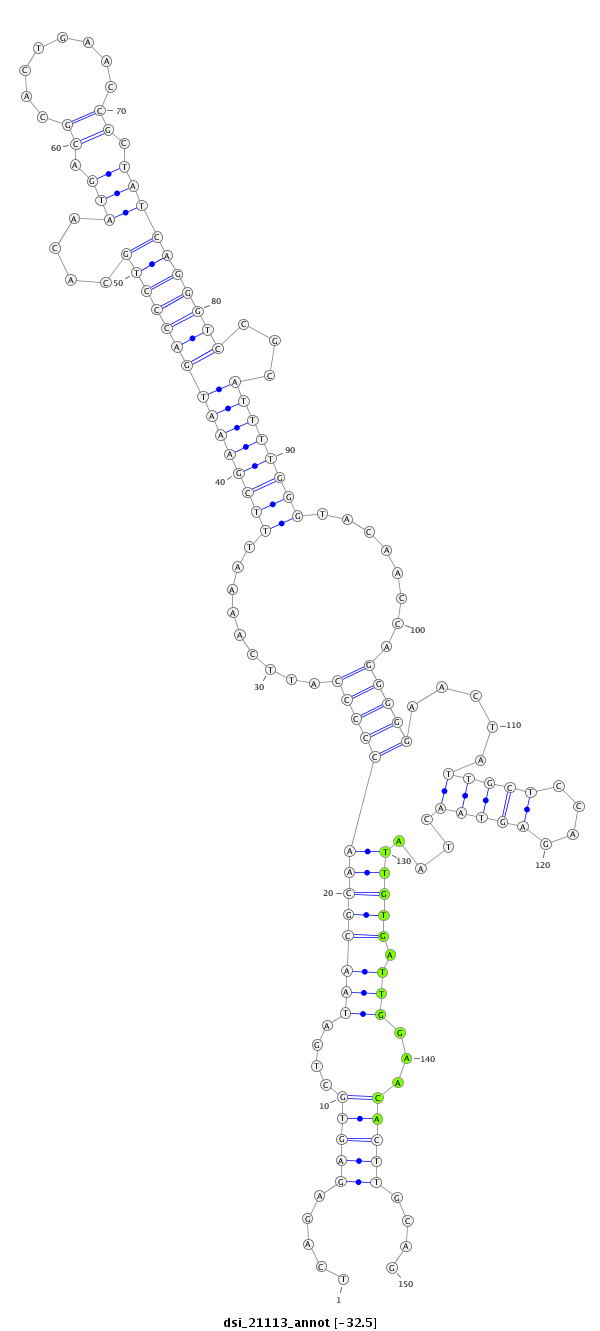
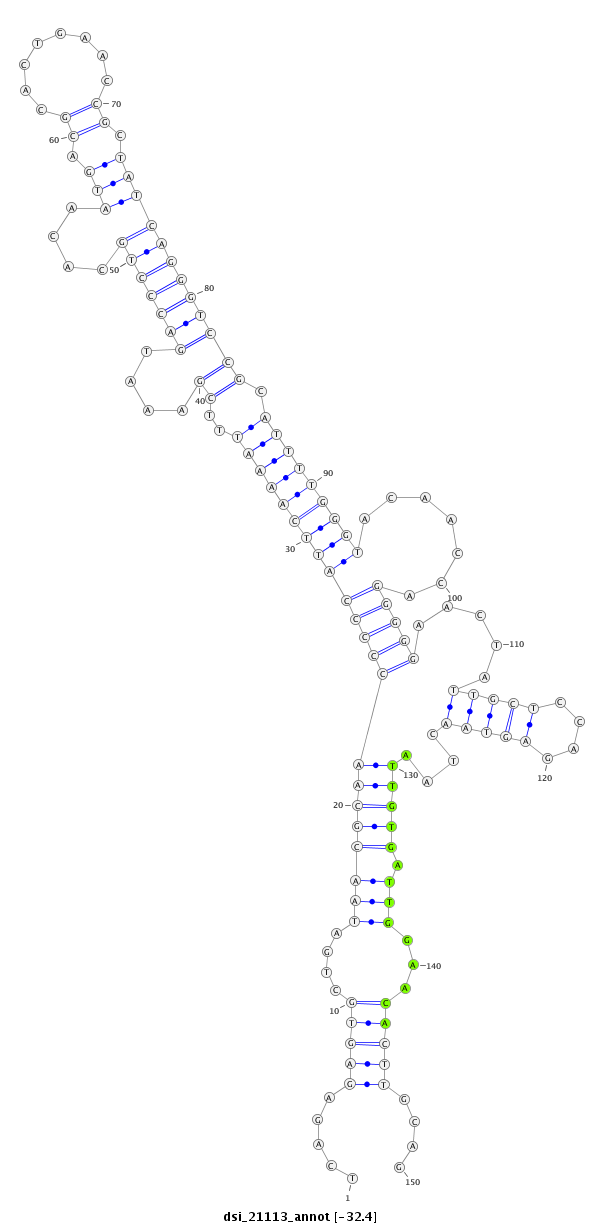
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -32.6 | -32.5 | -32.4 |
 |
 |
 |
CDS [2r_4794804_4794908_+]; exon [2r_4794804_4794908_+]; intron [2r_4793645_4794803_+]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTAGTCGTAATTAGCGCCAATTATTGCGATTATGACAAAGCTGGAATTAATCAGAGAGTGCTGATAACGCAACCCCCATTCAAAATTTCGAAATGACCCTGCACAATGACGCACTGAACCGCTATCAGGGTCCGCATTTTGGGTACAACCAGGGGGAACTATTGCTCCAGAGTAACTAATTGTGATTGGAACACTTGCAGAAACCGCGATGGGGCAACCTCCTGCTGCTTGTGGCCCTGGCAACCACATT **************************************************.....((((((((((..((((((((((.........(((((((((((((((....(((.((........)).))))))))))...))))))))........))))).....(((((....)))))....))))))))))..)))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
M053 female body |
SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
M024 male body |
M025 embryo |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................GTGCTGATAACGCGA.................................................................................................................................................................................. | 15 | 1 | 20 | 4.65 | 93 | 53 | 27 | 10 | 0 | 3 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................TCCTGCTGCTTGTGGCCCTGGC......... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................ATTGTGATTGGAACA......................................................... | 15 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CGCAGAAACAGCGATGGGG.................................... | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ACCAGGGCGAACTTTTGCT.................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TGTGATTGGAAGACT....................................................... | 15 | 1 | 12 | 0.17 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................CAGAAAGTGATGATGACGC.................................................................................................................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...................................................CAGAGAGTGATGTTGACGC.................................................................................................................................................................................... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...................................AAAGACTGGAATTAATCAGA................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................ACTGGAATTAAACAGAGACT............................................................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................ATCAGGCTGTGCTGATAA....................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................TGCACGATGACGCACC....................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................AGAGAGTGCTGATCCC...................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
AATCAGCATTAATCGCGGTTAATAACGCTAATACTGTTTCGACCTTAATTAGTCTCTCACGACTATTGCGTTGGGGGTAAGTTTTAAAGCTTTACTGGGACGTGTTACTGCGTGACTTGGCGATAGTCCCAGGCGTAAAACCCATGTTGGTCCCCCTTGATAACGAGGTCTCATTGATTAACACTAACCTTGTGAACGTCTTTGGCGCTACCCCGTTGGAGGACGACGAACACCGGGACCGTTGGTGTAA
**************************************************.....((((((((((..((((((((((.........(((((((((((((((....(((.((........)).))))))))))...))))))))........))))).....(((((....)))))....))))))))))..)))))....************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M024 male body |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................TCGCGGTAGTCCCAGGCGTA................................................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................ACGTGCTACTGCGTG........................................................................................................................................ | 15 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................TCCCAGGCGTGACAGCCAT......................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GGAGGATGACGAACA.................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................TTGGGGGCAAGTTTTAAG.................................................................................................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................................CTAAGCTTGTGAACGTA.................................................. | 17 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ........................................................................................................................CGATAGGCCCGGTCGTAAA............................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
Generated: 04/23/2015 at 04:55 AM