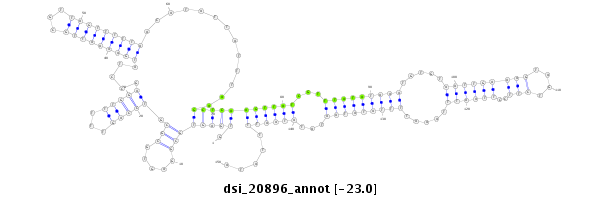
| droSim2 |
3r:678951-679200 + |
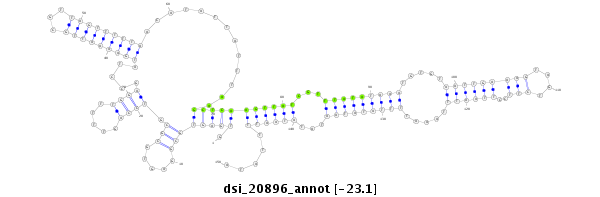
dsi_20896 |
ACAGC------------------------GCGGTCCCGATCCGGCTCGTGTGCATCCAGGCGATCACAATACAGGTGAGTGCGG-AGTGCGCGTCCA------GT-T-TTTGG---------------------------------------------------------------------------------------------------AGCCTATGAAAATTGC--------------GTTA---G-----TTT-------------------------------------TTAACATATTATTTAAGCTCAAGTTATCGTTTATAT---------------------------------------------------------------------------------------------------------------------------------------AA---------------ATGTG--T----AATTGAAAGTAGTCTTCC---TTAATTTAAATTTTATATAATATATAATTTTATATTTATAC--------------------------------------------------------------ATTGTAT-TAATTTAAGCGAATTATATTT-TAAAT------GAT---TACTTAA |
| droSec2 |
scaffold_6:848234-848485 + |
|
ACAGC------------------------GCGGTCCCGATCCGGCTCGTGTGCATCCAGGCGATCACAATACAGGTGAGTGCGG-AGTGCGCGTCCA------GT-T-TTTGG---------------------------------------------------------------------------------------------------AGCCTATGAAAATTGC--------------GTTA---G-----TTT-------------------------------------TTAACATATTATTTAAGCTCAAGTTATCGTTTATAT---------------------------------------------------------------------------------------------------------------------------------------AA---------------ATGTG--T----AATTGAAAGTAGTCTTCCT-GTTAATTTAAATTTTATATAATATATAAATTTATATTTATAC--------------------------------------------------------------ATTGTAT-TAATTTAAGCGAATTATATTT-TAAAT------GAT---TACTTAA |
| dm3 |
chr3R:752004-752243 + |
|
ACAGC------------------------GCGGTCCCGATCCGGCTCGTGTGCATCCAGGCGATCACAATACAGGTGAGTGCCC--------GTCCA------GT-T-TTTGG---------------------------------------------------------------------------------------------------AGCCTATGAATATTGC--------------GTTA---G-----TTT-------------------------------------TTAACATATTATTTAAGCTCAAGTTATCGTTTATAT---------------------------------------------------------------------------------------------------------------------------------------AA---------------ATGTG--T----AATTGAAAATAGTCTCCCCTCTTGGTTTAAGTTTTATATAATATATAATTTTA------TAC--------------------------------------------------------------ATTGTAT-TAATTTAAGCAACTTATATTT-TAAAT------GAT---TACTTTA |
| droEre2 |
scaffold_4770:1050268-1050504 + |
|
ACAGC------------------------GCGGTCCCGATCCGGCTCGTGTGCATCCAGGCGATCACAATTCAGGTGAGTCTCC--------GTCGA------AT-T-TACG-----------------------------------------------------------------------------------------------------GTCTATGACTATTGC--------------GTTA---G-----TTT-------------------------------------TTAACATATTATTTAAGTTCAAGTTATCGTTTATAT---------------------------------------------------------------------------------------------------------------------------------------GA---------------ATCTG--T----AATTGAAAGTAGTGTTTCT-CTCGTTACAAGTGTGATATAACATATAATTTAA------TAT--------------------------------------------------------------AATATAT-TCATTTAAGAGACTTATATTT-TAAGT------GAT---TACTTTA |
| droYak3 |
3R:1073468-1073706 + |
|
ACATC------------------------GCGGTCCCGATCAGGCTCGTGTGCATCTAGGCGATCACAATACAGGTGAGTGCCC--------GTCCA------GT-T-TAAGA---------------------------------------------------------------------------------------------------AGCCTATGAAAATGGC--------------GTTA---G-----TAT-------------------------------------TTAACATATTATTTAAGTTCAAT---TCGTTTATAA---------------------------------------------------------------------------------------------------------------------------------------AA---------------ATCTG--T----AATTGAAGGTAGTGTTTCC-CTTGATTTAAGTTTTAAATAAAATATAATTTAA------TAA--------------------------------------------------------------AATATGT-TAATTTAAGTGACTTATATTT-CAAAT------GATATATATTTAA |
| droEug1 |
scf7180000409759:702628-702841 + |
|
ACAGC------------------------GCGGTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCCCC--------CTCCG------AT-C-TCAGA-----------------------------------------------------------------------------------------TG-AGTTTTAAG-------------------------------------------------------TTCATTAAGTTTAT-TGTTAAATC----------------------A-------------------------------------------------------------------------------------------------------------------AATACTTTT-TGT------------T---------AT---------------ATGTG--T----AGAAGAGTG-AGTGCTCC---CTGACTCATATTTA---------------------------------------AGTTATTAGTAAT---------TAATTT--------------------AT------A-TAACATAAGCGGCATATATTT-TAAA-------------TACTTTA |
| droBia1 |
scf7180000302411:503080-503290 + |
|
ACAGC------------------------GCGGTCCCGATCCGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTAAGTCCCC--------GTCTGGGTCTGG-CA-TCTAGA----GATGCTAATGA------------------------------------------------------------------------------------ATTCAACATCT-------------TTTAC-T---------------------------------------------------------------------------TATATTTCATTTATACA----------------------------------------------------------------------------ATTCAATT-------------------------------------------------------------------AATAACATACTGGAGCGTTGTGTTCC---CGGACTCGCATTTA---------------------------------------AGTTA--------TGTAGTGAA-----------------------------------TTAC-C-------ATTTTACTT-TGAAT------AA--------GAA |
| droTak1 |
scf7180000415789:448903-449121 + |
|
ACAGC------------------------GCGGTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCCCC--------GTCCG------AA-T-ATATG---------------------------------------------------------------------------------------------------AACTTAAGTTAT-------------TTTGCGT---------------------------------------------------------------------------TATCGTTAATTTATATA-----------------------------------------------------------------------------------------------AATACCTCTATGT------------T-------------------TTTACAACACA-----CATACTGTA--GTGGTGTTTCC-CTTGATTATCATTTA---------------------------------------AGCTA--------TGTAGTAAA-----------------------------------TTAATCAGAACGACTTATATTT-TAAAT------AAA---TAC-TAA |
| droEle1 |
scf7180000491104:2546183-2546443 + |
|
ACAGC------------------------GCGCTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTAAGTCTCC-AGTTTG-------------T-T-TCAG---GATTTCCTCAAGGATTTTTGAGTTAGTTTTGTACTATAATTTTTACTGTTAAGTTACCGTACAATCA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTCAAT----------------------------------------------------------------------------AGTCATTTCATGATTGTTCTTATAAAAGTTATC-TTA-------------------ATACCTAGTACTCC--CATGATTCACATTTC---------------------------------------AGTTA--------CAAATTAA------------------------------------ATAATCTAAGAGACTTATATTT-TTAAT------GAT---TACATTA |
| droRho1 |
scf7180000772422:19745-20005 - |
|
ACAGC------------------------GCGTTCCCGATCACTCTTGTGTGCCTCCAGGCGATCCCAATTTAAGTGAGTCCTT-AGTGTG-------------T-T-TTAGA-TAAAACTTTTAATGATTTTTAAGTTAGTTATGT------------------------------------------------------------------------------------------------------------------------------------------------------ACTATAATTTTTGCGTTAAAGTCGTCGTACACTTA---A------------------------TTTCTAT----------------------------------------------------------------------------GGTGAATTTATGATTGTTCTTATAAAAGTTATTTTTA-------------------GTACCCAGTGCTCCC-TCTGATTCAGATTTC---------------------------------------AGTTA--------CATAT----------------------------------------CAATTTAAGAATCTTATATTT-TA-AT------GAT---TACTTTT |
| droFic1 |
scf7180000453850:105058-105254 + |
|
ACAGC------------------------GCGGTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCCCC-CGACTG-------------T-C-CTCCA-TGAATGTGCTAATGATTTTTAATTTAACTGAGAACTT---------------------------------------------------------------------------------------------TA---G-----T-------------------------------------------------TTTATCGTTTAAGTTATCGTTAAGTTA---A------------------------GTT------------------------------------------------------------------------------------------------------------------------ACCTG--A----AAA--------------------------------------------------------------------------------------------------------------------------------TGAGTCTAAGAGGCTTATATTA-TATTTGTAGACGAT---TACTTTT |
| droKik1 |
scf7180000302634:451897-452132 + |
|
ACAGC------------------------GCGCTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCTTC-C---------ATTA-----------TG-----------------------------------------------------------------------------------------------------TTTTTTAACTATTAT--------------GTTACCAATTTCATC------------------AAGATG----CTTAATATAATTTTTGTA-CAATTAAGA--TAT---TTTTAGTAAT-----AATTTAA---TATATAAATATAATCTT-TTTATTGAATGTCTGTTTATAGTCTTGTCTGGTTATACTTTAATTTCATTGAAATTATTCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AA----------------------TTTTGA |
| droAna3 |
scaffold_13340:22963662-22963901 + |
|
GCAGC------------------------GCGCTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCACT-C---------AGGATCCTT-CAA-CTGGA--------------------------------------------------------------CTATCATATCCTTCTTAAATTTATTCTATGTTTTTATG----AGATTTTT----------A--------------------------------TT--------------CTCAGTTTTA--TTCATACTTTTTTAAC-----------------------------------------------------------------------------------------------------GAAATTATTCAAAGAGGAATAGAATT--------------------------TG---------------T------------------------------------------------------------TTTGTGATAAATAGA--------------------------------ATAGTTATAATTTAGTTAAAGTG-------------------------ATT----AT-GTTAA------TAC---TACTTAA |
| droBip1 |
scf7180000396424:1231607-1231836 + |
|
CCAGC------------------------GCGCTCCCGATCAGGCTCGTGTGCATCCAGGCGATCACAATTTAGGTAAGTCAGT-C---------AGGATCCTT-CAA-CTGCA----G----------------------------------------------------------------------------------------------ATCCATGATTTGTTTAGATTTCAA--------------------------------TT----------------------------CATTTTATTTTA-TTCAAT---TT--TTATTA---------------------------------------------------------------------------------------------------------------------------------------AA---------------ATTTAAAT----ACAA---AAGGGTGCTTAT-AAGGGTACCATTTTTAAAGAAAACTTA------------TCA----------------------------------ATAGGTTTAG------------------------T-TTAGTTGAATGAATT----TT-GTTAA------TAT---TATTTAA |
| dp5 |
2:22041305-22041570 - |
|
TCAGCCAC------AGCCACCGCCACGCAACGCTCCCGATCAGGCTCGTGCGCATCCAGGCGATCACAATTTAGGTGAGTGCTGCC---------ATG------G-C-ACAG---GAGTGC---------------------------------------------------------ATCAACCTCTACCTCCATTTAAGTTG-GTTCTTAAGCCCAAGATTATT-----------------------------------------------------------GTTAATTATATTTTCATATGATTTAAGC--GAT---TCCTAGAGAT---------------------------------------------------------------------------------------------------------------------------------------AA---------------T------------------------------------------------------------TTTGTAATTATACAA--------------------------------TGATTTGTAAACCATTTAAGGTGTGGAT------T-TAACATCCGCGCAATAGATTT-TATAA------TAT---GATTTAC |
| droPer2 |
scaffold_3:4822739-4823005 - |
|
TCAGCCACAGCCACAGCCACCGCCACGCAACGCTCCCGATCAGGCTCGTGCGCATCCAGGCGATCACAATTTAGGTGAGTGCTGCC---------A-------------CAG---GAGTGC---------------------------------------------------------ATCAACCTCTACCTCCATTTAAGTTG-GTTCTTAAGCCCAAGATTATT-----------------------------------------------------------GTTAATTATATTTTCATATGATTTAAGC--GAT---TCCTAGAGAT---------------------------------------------------------------------------------------------------------------------------------------AA---------------T------------------------------------------------------------TTTGTAATTATACAA--------------------------------TGATTTCTAAACCATTTAAGGTGTGGAT------T-TAACATCCGCGCAATAGATTT-TATAA------TAT---GATTTAC |
| droWil2 |
scf2_1100000004943:8448086-8448289 - |
|
ACAGC------------------------GAGGTCCCGATCCGGGTCGTGTGCATCCAGGCGATCACAATTTAGGTGAGTCTGT-C---------AAT-TCGA-TCCA-CTCAA--------------------------------------------------------------ATT-------------------------------TAAT-------------------------------------------------------TTCATTAAGTTAATTTGTTAATTTTATGAT-------------------------------------------------------------------------------------------------------------------TAAATT-TTAAAAATGGAAAACATTT-CGAAC-------------------------------------ATTTG--T----GGGA-AGTG-A----------------------------------------------------GATCATTTTCCAGTTATTATTAAT---------T-----------------------------------------------CTT----AT-TTAAA------TGG---TAATTAA |
| droVir3 |
scaffold_12855:8525851-8525906 + |
|
ACTGC------------------------GCGCTCCCGATCTGGGTCGTGCGCATCAAGGCGATCACAATTTAGGTAAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6540:23725281-23725484 + |
|
ACAGC------------------------GCGCTCCCGATCTGGGTCGTGCGCATCAAGGCGATCACAATTTAGGTAAGCGCGC-C---------AAA------T-C-A------------------------------------------------------------------------------------------------------------------ATTGA--------------GATT---A-----TTT----------------------------ATAATATAACT------TTGTATAGTTTTTAGTTATCGTTATTGC---------------------------------------------------------------------------------------------------------------------------------------AA---------------C------------------------------------------------------------TTTTTAGCTAATCTA--------------------------------TGATTTAGATTAAGCTCTATATTTATTCA-------TGACTTAAGCCTAATTTAGTTTTAGAT-----CTAT---TATTTAA |
| droGri2 |
scaffold_15074:5107778-5108014 + |
|
ACAGC------------------------GCGCTCCCGATCTGGCTCGTGCGCATCAAGGCGATCACAATATAGGTGAGTCGTT-T----------------------TTTGT---------------------------------------------------------------------------------------------------TTTCCATGAATATTAC--------------GACTCCAGTTTCATCTATAATATATTT-TTGTTAAGTTTATATTTTGGGAC----------------------A-------------AT-----AACTTGATAGTATATAAAATGAAGCTTCATC----------------------------------------------------------------------------AATAATTCTGTGATTTTTGTTACAA-------------------------------------------------CAATCTGCATTT--------------------------------------------------------------------------------------TGAT------T-AAAAATC--------AAGCTT-AATAA------TAG--TAATTTAA |