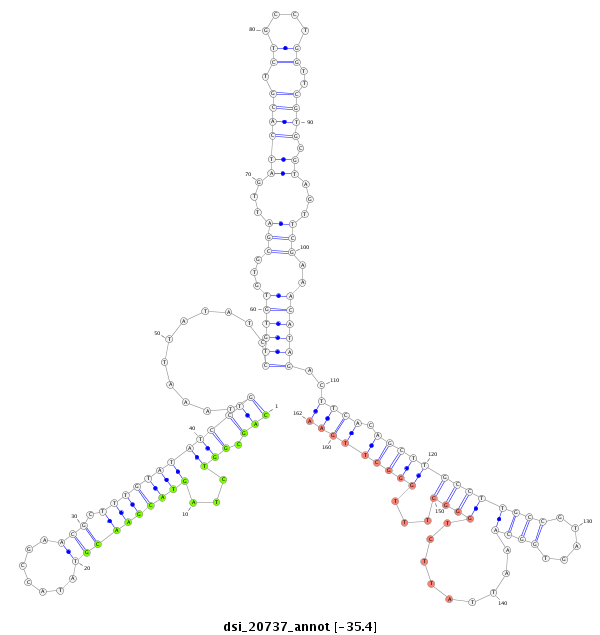
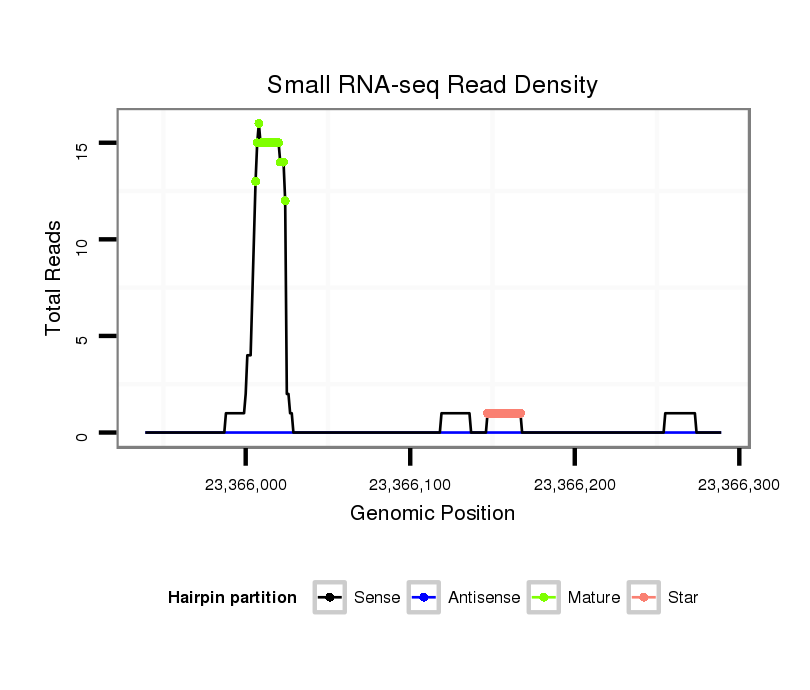
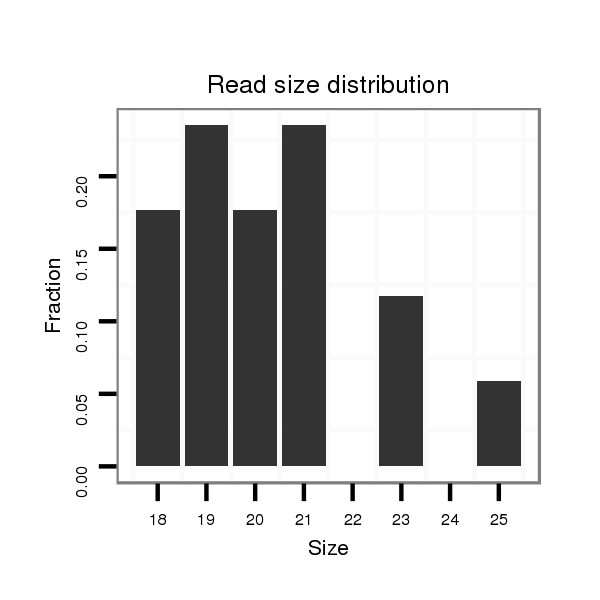
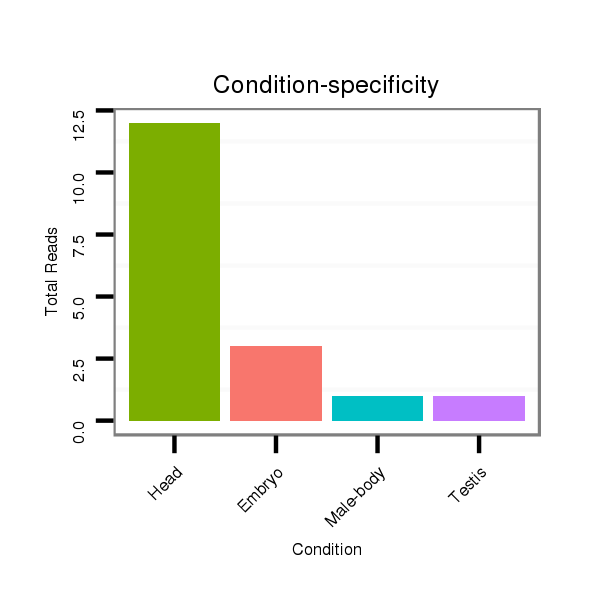
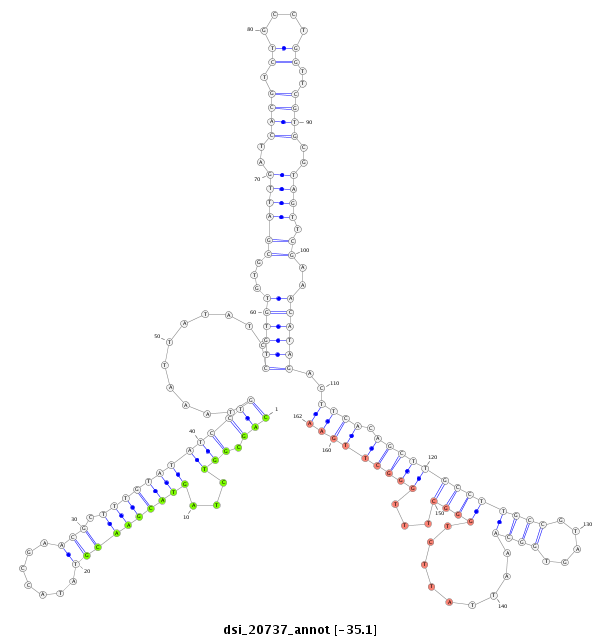
ID:dsi_20737 |
Coordinate:3r:23365989-23366239 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
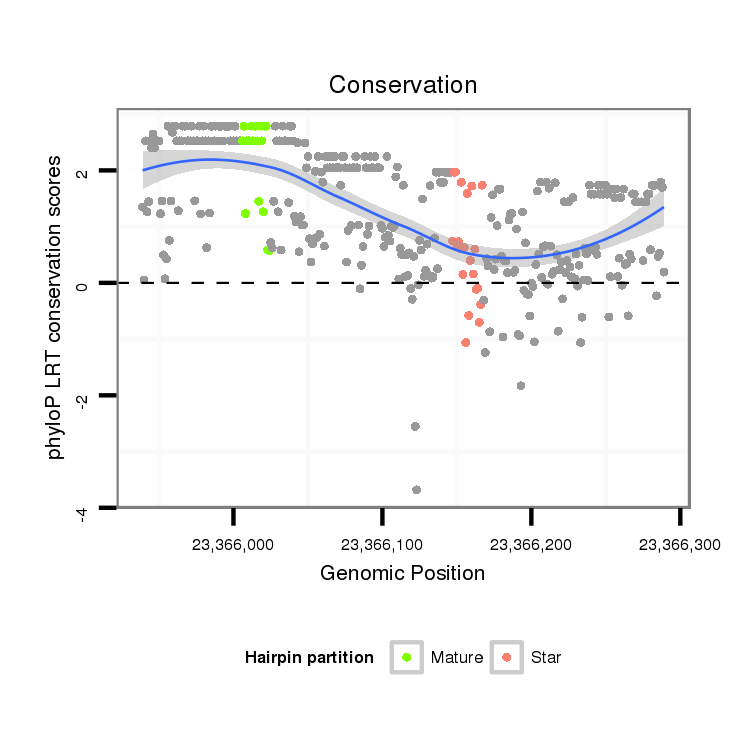
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -35.1 | -35.1 | -35.1 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TGGCAGCAGCGCCTCCGAATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATACCGAACGCTTTGTATATCCTGTAAATTATATCCTGTGTGTGCGATTGATCACGTCTGCCTGGTTCGTGCGTAGTTCGAAACATAGACTTCACAGCTTGCCTTGCCGTAGTGGCAAATTATTCTGGGCTTTGGGCTTGAACCTTATATTTTTTTAAAGATCACCTCAACCCATACACGACATTTATCAAGCTGGTCTTATCGCGCACTTGGAGTCTCTGACTTCTAAAGTTGCCAGCAAATTCGCGCGCGACAAAAACTCAG
*******************************************************************(((.(((...((((((((((.......))).)))))))))))))...........((((((...(((...((((((.((....))..)))).))...)))..))))))..((((.(((((((((((((.....)))).........))))...)))))))))************************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553487 NRT_0-2 hours eggs |
M025 embryo |
M024 male body |
O001 Testis |
O002 Head |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR902009 testis |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................CAGCGGTCTAGTACGAACG......................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CCAGCGGTCTAGTACGAACG......................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................TCGCCCAGCGGTCTAGTACGAAC.......................................................................................................................................................................................................................................................................... | 23 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................AGCGGTCTAGTACGAACG......................................................................................................................................................................................................................................................................... | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TGCGCGATTGATCACGTCGTC........................................................................................................................................................................................................... | 21 | 3 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CCCAGCGGTCTAGTACGAACG......................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CACGTCTGCCTGGGT..................................................................................................................................................................................................... | 15 | 1 | 5 | 1.40 | 7 | 0 | 5 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................AGTTGCCAGCAAATTCGCG................ | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................GCGGTCTAGTACGAACGTA....................................................................................................................................................................................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................ATTCTGGGCTTTGGGCTTGAA.......................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................GTGTGCTCCTGACGT................................................................................................................................................................................................................................................................................................................ | 15 | 1 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................GTCGCCCAGCGGTCTAGTACG............................................................................................................................................................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ACAGCTTGCCTTGCCGTA......................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................AGTAAGAAAAGCGTCGCCCAG......................................................................................................................................................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................TCTGGGCTTTGGGCTTGAACCG....................................................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................CCCAGCGGTCTAGTACGAACGTATA..................................................................................................................................................................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................AAAGATCACCTCATGCCAGA........................................................................................ | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................GCACGTCTGCCTGGGT..................................................................................................................................................................................................... | 16 | 2 | 20 | 0.25 | 5 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................ATGCGTGCGCTCCTGTCTTT............................................................................................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................TCGTCCAGCGGTCTAGG................................................................................................................................................................................................................................................................................ | 17 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................ATAGACCTGACAGCGTGCC................................................................................................................................................................. | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................GCGTCGCCAAGCGGT..................................................................................................................................................................................................................................................................................... | 15 | 1 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................GTCTAGCGGTCTAGGACG............................................................................................................................................................................................................................................................................. | 18 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............TCGAATTTTGGTCAATTCGT............................................................................................................................................................................................................................................................................................................................. | 20 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................AATTTTGGCCAATGA............................................................................................................................................................................................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
|
ACCGTCGTCGCGGAGGCTTAAAACCGGTTACGCACACGAGGACCGCAAGTCATTCTTTTCGCAGCGGGTCGCCAGATCATGCTTGCATATGGCTTGCGAAACATATAGGACATTTAATATAGGACACACACGCTAACTAGTGCAGACGGACCAAGCACGCATCAAGCTTTGTATCTGAAGTGTCGAACGGAACGGCATCACCGTTTAATAAGACCCGAAACCCGAACTTGGAATATAAAAAAATTTCTAGTGGAGTTGGGTATGTGCTGTAAATAGTTCGACCAGAATAGCGCGTGAACCTCAGAGACTGAAGATTTCAACGGTCGTTTAAGCGCGCGCTGTTTTTGAGTC
**************************************************************************************************************************(((.(((...((((((((((.......))).)))))))))))))...........((((((...(((...((((((.((....))..)))).))...)))..))))))..((((.(((((((((((((.....)))).........))))...)))))))))******************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553488 RT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR553485 Chicharo_3 day-old ovaries |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................................................CAGAGACTGAGGATT................................... | 15 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................TTGGGCCAGAATAGCGAGT........................................................ | 19 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................ATGCTTCGTATCGGAAGTG......................................................................................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................................................TCAATAGTTCGACCAGAT................................................................ | 18 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| ............GAGGCATAAAACCGGT................................................................................................................................................................................................................................................................................................................................... | 16 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................GGACCGCATCACCGTTT................................................................................................................................................. | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................................................ACGCTAGCGAGTGCTGACG........................................................................................................................................................................................................... | 19 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................GGAGTTAGGTATGTTCT................................................................................... | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| .................................ACACGAGGACAGCAA............................................................................................................................................................................................................................................................................................................... | 15 | 1 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................AACCTCAGAGACGGATGTT.................................... | 19 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................AGAAATTTCTAGTGG.................................................................................................. | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................TCACTAGTGCAGACCG.......................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:23365939-23366289 + | dsi_20737 | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCACGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCACA----------------G-CTTG---CCT----------T-GCC--GTAGTGGCAAATTATTCTGGGCTTTGGGC-TTGAAC----------------------------CTTAT---------------------------------A---TT-----------TT--T------TTAAAGATCA------C-----CTCAACCCATACA-----------------------------------------------------------------------------------------------------------------------------------CGACATTTATCAAG--------CTGGTCTT----------------ATC----G-----CGC-ACTT-GGAGTCTCTGACTT-CTAAAGTTGCCAG-CAAATTCGCGCGCGACAAAAAC----------------TCA---------G |
| droSec2 | scaffold_22:591276-591627 + | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGACCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCACGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCACA----------------G-TTTG---CCT----------T-GCC--GTAGTGGCAAATTATTCTGGGCTTTGGGC-TTGAAC----------------------------CTTAT---------------------------------A---TA---------T-TT--T------TTAAAGATCA------C-----CTTAACCCATACA-----------------------------------------------------------------------------------------------------------------------------------CGACATTTATCAAG--------CTGGTCTT----------------ATC----G-----CGC-CCTT-GGAGTCACTGACTT-CTAAAGTTGCCAG-CAAATTCGCGCGCGACAAAAAC----------------TCA---------G | |
| dm3 | chr3R:23972540-23972894 + | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCACGTCGGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCACA----------------C-CTTG---CCT----------T-GCC--GTAGTGGCAAATTATTCTGGGCTTTGGGC-TAGAAC----------------------------CT---------------------------------ATTA---T------------TT--T------TTAAAGATCA------CTCAATCTTAACCTATACA-----------------------------------------------------------------------------------------------------------------------------------CGACATTTATTAAG--------CTGGTCTT----------------ATC----A-----CGC-ACTT-GGAGTCAGTGACTT-CTAAAGTTGCCAG-CAAATTCGCGCGCGACAAAAAC----------------TCA---------G | |
| droEre2 | scaffold_4820:4067421-4067774 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCACGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCACA----------------G-ATTG---CCT----------T-GCC--GTAGTGGCAAATTATTCTGGGCTTGGGGC-TTGAAC----------------------------TT---------------------------------ATTATTATT-----------TT--T------TTAAAGATCA------C-----CTAAGCCTGAACA-----------------------------------------------------------------------------------------------------------------------------------CGAGATTTATGCAG--------CTGGTCTT----------------ATC----A-----CGC-ACTT-GGAGTCACTGACTT-CCAAAGTTGCCAG-CAGATTCGCGCGCGACAAAAAC----------------TCA---------G | |
| droYak3 | 3R:22522017-22522358 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCACGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCACA----------------A-CTCG---CCT----------T-GCCTAGTAGTGGCAAATTATTCTGGGCCTGGGAC-TTGAAC----------------------------TTTTT---------------------------------A---AT-----------TT--T----GCCAAAAGATCA------C-----CTAAACCTAAATA-----------------------------------------------------------------------------------------------------------------------------------CGATATTT--------------------------------------ATC----A-----CGC-ACTTGGGAGTCACTGACTT-CGAAAGTCGCCAG-CAAATTCGCGCGCGACAAAAAC----------------TCA---------G | |
| droEug1 | scf7180000409787:1626228-1626601 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCATA----------------C-ATTG---CCT----------T-GCCTTGCAATGGCAAATTATTCTGGTCTT-GGGC-TAACAC----------------------------CT-TT-----------------------------------T-TT----------T-TTGTATTG---AAACGATCCT-----C-----TGTA--------GC-AACCT-TCATTT-----AT------------------------------------------------------------------------TTGTAT---------------------------TTCTGCGACTTTTATTAAG--------TTGGTCTT----------------ATC----A-----CGC-CTTT-GGAGTCACTGACTT-CTAAAGTTTCCAG-CAAATTCGCGCGCGACAAAAAC----------------CCA---------G | |
| droBia1 | scf7180000302075:2835718-2836094 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTGTAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTGC--CTGGTTC--GTGTTCGTAGTTCGCAGCATAGACTTCTTT------------CT--G-ATTGCCGCCT----------T-GCT--GTAATGGCAAATTATTCTGGGCCT-GGGC-TTAAATGCAATATAT---------ATATATAAT-AT-AT---------------------------------A---TT-----------TT--T------TTTACGATCA------C-----CCCA--------GCC-----TGAGTTAACTTAATTTAT-----------------------------------------------------------------------------------------------------TTG------------TA-------------A------------AC-------CC----ACAACTCGTTG-CT-GGAGTCACTGACTT-CTTAAGTTGCCAGCACAATTCGCGCGCGACAAAAAG----------------CCA---------G | |
| droTak1 | scf7180000415383:555856-556219 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGATACGATTGATCTCGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCTTT------------CTGAGATTTG---CCC----------T-GCT-TGTAATGGCAAATTATTCTGGGCC-------ATGAACAC--------------------------AT-AT---------------------------------A---AT-----------TT--T------TAAACGATCA------C-----AGCA--------GCCA-CCTTTAGTT------ATTTAT-----------------------------------------------------------------------------------------------------TT------------TTAAG--------CTGGTCCT----------------ATCGCAAA-----TAC-TTTT-GGAGTCACTGACTT-CTAAAGTTGCCAC-CGAATTCGCGCGCGACAAAAAG----------------CCA---------G | |
| droEle1 | scf7180000491280:4028992-4029324 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TT---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATAGACTTCTT-------------CTGAC-TC------------------T-GCCT-CTAATGGCAAATTATTCTGGGCT---GGC-TCACAC----------------------------AC-TT---------------------------------A---TT-----------TTTGT--TGGTTAAACGATCAT-----------------------G----CC-TTAGTTT-----TT-----G------------------------------------------------------------------TTATAT---------------------------TTGTACAACTTT----------------------------------------------------CAC-ATTT-GAAGTCACTGACTTTCT-------------GAATTCGCGCTCGACAAAACC----------------CCA---------G | |
| droRho1 | scf7180000779538:271585-271950 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA-TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGGTCTCGTCTGC--CTGGTTC--GTG--CGTAGTTCGAAACATTGACTTCT------------------G-ATTG---CCT-----------------CTAGTGGCAAATTATTCTGGGCTT-GGGC-TTGCA---------------------------------T---------------------------------A---TT---------TT-TTGT------TTAGCGATCATGA-----------------GATGGC-AGCAT-A---------------------------------------CGTTTAAGTTTCC--------------------------------TAGCAT---------------------------TTTCACAACTTTCACTGAG--------GTGA------------AT-------CC----A-----CGC-ACTC-ACAGTCACTGACTT-CTGAAGTTGCCAGCCGAATTCGCGCGCGACAAAAAC---------------CCCA---------A | |
| droFic1 | scf7180000454021:45231-45600 - | TGGC------------------------------AGCAGCGCCTC--CG-AATTTTGGCCAATGCGTGTGCTCCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAA--TTA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTGC--CTGGTTC--GTG--CGAAGTTCGAAACAAAGACTT-----------------------TTG----CT----------T-GCC--GTAATGGCAAATTATTCTGGGCCT-GGGC-TTACAC--------------------------------T---------------------------------A---TT-----------TT--T------TTAACGATTATGATCACTCAC-AGCA--------GCCAGCC-CTGGTTT-----GC------------------------------------------------------------------------TTATAC---------------------------TCACACGACTCTAATTAAG--------TTGGTCTT----------------ATCCC--A-----CGC-ATTT-GCAGTCACTGACTT-CCAAAGTCGCAAGGCAAATTCGCGCGCGACAAAATC---------------CCCA---------G | |
| droKik1 | scf7180000302475:843849-844269 - | TGGC------------------------------AGCAGCGCCTC--TG-AATTTTGGCCAATGCGTGTGCTGCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAAAATA---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTGC--CTGGCTC--GTG--CGTTGTTCGAAACACAGACTCCTAA----------------C-TCTG----AT----------T-GCA--ATAATGGCAAATTATTCTGGTCTT-AGGA-----AG-CC-------------------------CT--------------CAT----GC-C---GTTCTATTATTTT--T---TTGTTT-TTGT------TTAA------------------ATTAACATGATCGC-A-----------------------------GATTTGTCTAG-----------AGTCTGT--------------------------------TTGCATCTTCATC----------GCTAAACTTT---GGCGACTTTCATTAAACTTTGGCGCCGGTGTTTGCGAACGAT-------TC----T-----CGC-AC-T-GCAGCCACTGACTT-CTAAAGTTGCGA-----------------CGAAACCAA-----------AAGCCATCATCAGTGG | |
| droAna3 | scaffold_13340:22115098-22115590 + | TGGC------------------AGTGGCTTCGAAAGCAGCGCCTC--TG-AATTTTGGCCAATGCGTGTGCTGCTGGCTTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTAT--ATCCTGTAAA---A---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTAT--TTGGTTC--GTG--CGTTATTCGAGACACAAAGGACACAAAGA--------------TTTTCTTCCT----------C-TCC--ATAATGGCAAATTGTTCTGGGACT-GGGA-ATAAATTCA------------------AATAA-A-----ATTGTAGACAGCGTATATCAATTTT----------T-TTTTTCGTTA---------------------------------------------------------------------TTTTTTGCCAAAGATCA-TC-CGATCATCGTTTAAGCTTGT--------------------------------TTGTAT-TTCACCCTTGGACGGCTGCAAACTTT---GGCGACTTTCATTAAACTTTGGCA--AATGTTTGCAGATGATATTCGCGTCAG--A-----C---G-CT-GCAGTCACTGACTT-CTAAAGTTGCCAG-CGATTTCGCGCGCGACAAAAACAAAGCCAAAGCTAAAGCC--CATCAGTGG | |
| droBip1 | scf7180000396583:59448-59936 - | TGGC------------------AGTGGCTTCGAAAGCAGCGCCTC--TG-AATTTTGGCCAATGCGTGTGCTGCTGGCTTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCCTTGTAT--ATCCTGTAAA---A---TA---------------------------------------TC-----------------------CTGTGTGTGCGAT-----TGATCTCGTCTCT--TTGGTTC--GTG--CGTTATTTGAGACACAAAGGACACAAAGA--------------ATTGCTTCCT----------T-TCC--ATAATGGCAAATTATTCTGGGACT-GGGAAATAAATTAA------------------AATCA-G-----ATTATAGACATCGTATATAAATTTTC--------------TTCGTTGTTT-TTTT------T------------------------------------------------------------CTCAAAGATCTGTCTCGATCATCGTTTAAGCTTGT--------------------------------TTGTAT-TTCACCCTTGGTCGGCTGCAAACTTT---GGCGACTTTCATTAAACTTTGGCA--AATGTTTGCAGATGATTTTCGCGTCAG--A-----C---G-CT-GCAGTCACTGACTT-CAAAAGTTGCCAG-CGAATTCGCGCGCGACAAAAACAAAGTCA------AAGCC--CATCAGTGG | |
| dp5 | 2:8107590-8108089 - | TTGCCGTTGCAGTGGCAGTGGCTGTGGCTGTGGCAGCAGCGCCTC--TGAATTTTTGGCCAATGCGTGTGCTGCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTGTTCTTCCTGTTCT-TTGGGTT--CCACCTCCGTTA---CCGTATAGACGA-----CGTATATCTGAAGAGAAGAGATACGAGTGGTGTGTGTGTGCG-T-----TGATCTCGTCTCTTTTTGGTTCCGGTG--CGTTACTCCAAA------AGAAACAGAAAATCTGAAACTGCGAACTG---CTCTGAAAAGCTTTAACC--ATAATGGCAAATTATTTGTGGCG---GAAAGTG----CTCTACAAATGATCTATTGAGATCA---T---ATCAT-GAGATCACGCCTGC-CTTACTTCTATTATTAT--T---CTG--------------------------A------------------------------------------------------------TCCAT-----------AGTCTGTGATGTCTTTAAGCCCAGTTTCCCAGTGAGGGCTTCCACT----------GTCAGCCACAAACTTTTTGGGCGACTTTCATTAAACTTTGA--------------------------------------------------------------------------------------------------------------------CA---------A | |
| droPer2 | scaffold_0:1942018-1942153 - | TTGCCGTTGCAGTGGCAGTGGCTGTGGCTGTGGCAGCAGCGCCTC--TGAATTTTTGGCCAATGCGTGTGCTGCTGGCGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACGAACGTATAC--CGAACGCTTTGTGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004921:1209746-1209876 + | TTGTCGT------------CGCTGTCGCTGAAGAAGCAGCGCCTTTTC--AAATTTGGCCAATGCGTGTGCTGCTGACGTTCAGTAAGAAAAGCGTCGCCCAGCGGTCTAGTACAAACGAGTACTCTGTACGCTTTGTGT--TCCTT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_13047:15568591-15568813 - | CAGC------------------------------GGCAGCGCTTT--TC-AAT-TTGGCCAATGCGTGTGCTGCTGACGTTCAGTAAGAAAAGCGTCGCCCAACGGTCTAGAACGAACCCGAAC--CGTACGCTTTGAGTTGATTCTGTTGT-TTGGAGTAGCCAGCGACGATATAGTCGTAAAGTCGAACAGTCGTATA-------------GATATATAAAGAGTGTGTGTGCG-T-----TGCGCTCGTCT-C--TTGATTC--GTG--CGTTACTCGAAACA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_1756:5262-5364 + | CGGC------------------------------AGCAGCGCTTAT-CA-AATTTTGGCCAATGCGTGCGCTGCTGGCGTTCAGTAAGAAAAGCGTCGCCCAACGGTCTAGTACGAAATCGTAC--CGTACGCTTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14830:4421176-4421276 + | CAGC------------------------------AG---CGCTCTTGCA-ATTTTTGGCCAATGCGTGTGCTGCTGGCGTTCAGTAAGAAAAGCGTCGCCCAACGGTCTAGTACGAAAGCGTAC--CGCACGCTTTG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 11:07 PM