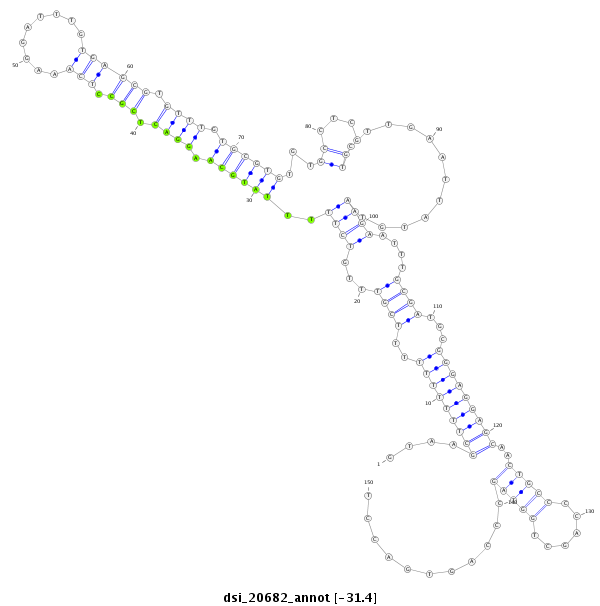
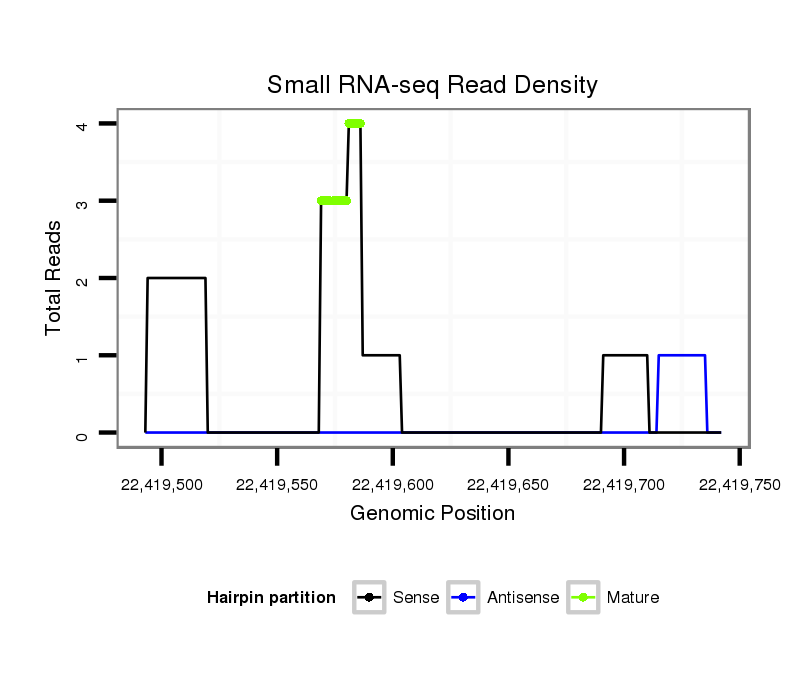
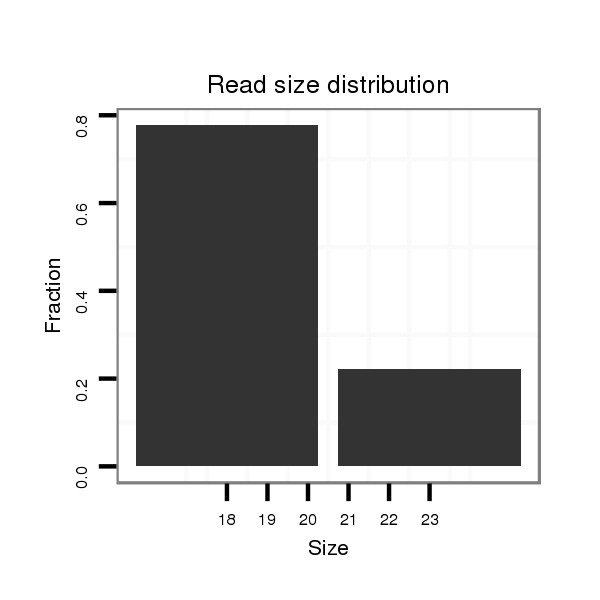
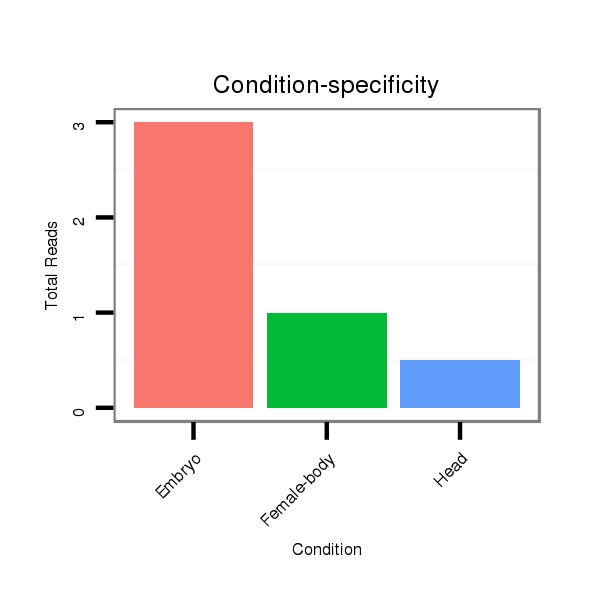
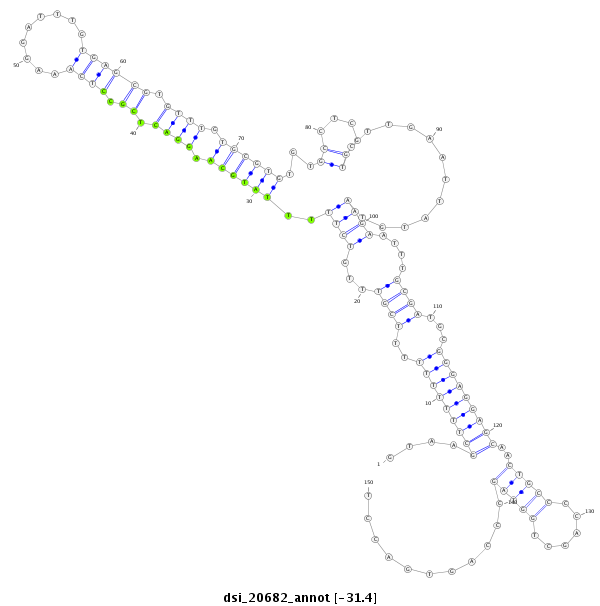
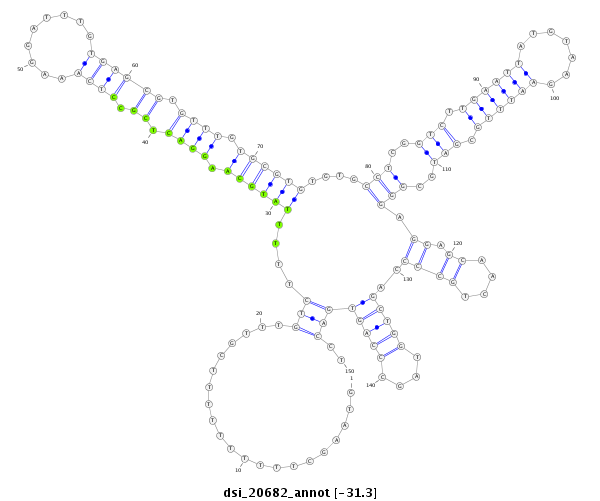
ID:dsi_20682 |
Coordinate:3r:22419543-22419692 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
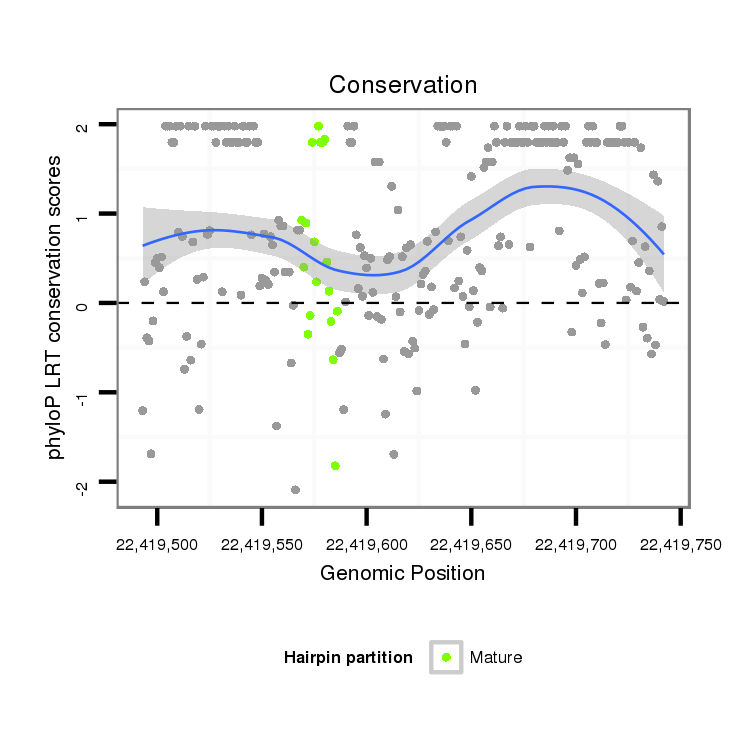
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.4 | -31.3 | -31.3 |
 |
 |
 |
CDS [3r_22419526_22419542_+]; five_prime_UTR [3r_22419282_22419525_+]; exon [3r_22419282_22419542_+]; intron [3r_22419543_22421784_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAGCGCAACGGAAAGCAATTATACGACATAAAAATGTTCTCACGACCCAGGTAAGCTTTTTTTTTTCGTTTGTCTTTTTATGCAAGGACTCGCCTCAAAGGATTTGTGAGCGTGTTTGTGCGTGTGTGCCTCGGTCTTGAATTATGTAAGAATTTGCGATGCGGGAGGAGCAACTGCCCCAGCTGGTAGCCCAGTGACCTAGTCACTCGGATCAGACGAGCGGCCGCCAAGGCGTCCGTGTATATATACA **************************************************....(((((((((..((((...((((..((((((.((((.(((.(((.........)))))).)))).))))))...((....))............))))....))))...)))))))))..(((((......)))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M024 male body |
SRR553487 NRT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
M053 female body |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
SRR553485 Chicharo_3 day-old ovaries |
M023 head |
SRR1275487 Male larvae |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
O001 Testis |
SRR902008 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................AGAACTGGCGATGCGGGA.................................................................................... | 18 | 2 | 7 | 7.00 | 49 | 0 | 11 | 20 | 5 | 6 | 0 | 2 | 0 | 1 | 2 | 0 | 1 | 1 |
| ............................................................................TTTATGCAAGGACTCGCC............................................................................................................................................................ | 18 | 0 | 1 | 3.00 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGCGCAACGGAAAGCAATTATACGAC............................................................................................................................................................................................................................... | 26 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GAATTGGCGATGCGGGA.................................................................................... | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................TTTTTATGCAAGGACTCGCCC........................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AACTTGCGATGCGGGA.................................................................................... | 16 | 1 | 2 | 1.00 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CTCAGATCAGACGAG.............................. | 15 | 1 | 3 | 1.00 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAATTGGCGATGCGGGA.................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................TTTATGCAAGGACTCGCCC........................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................CTAGTCACTCGGATCAGACG................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AATTGGCGATGCGGGA.................................................................................... | 16 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAACTGGCGATGCGGGAGG.................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................CTCGCCTCAAAGGATTTGTGAGC........................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAACTGGCGATGCGGGAGGA................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAATTGGCGATGCGGGATG.................................................................................. | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAACTGGCGATGCGGG..................................................................................... | 17 | 2 | 20 | 0.90 | 18 | 0 | 10 | 5 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................AATTGGCGATGCGGGATG.................................................................................. | 18 | 2 | 5 | 0.60 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CCTCAGATCAGACGAG.............................. | 16 | 2 | 20 | 0.55 | 11 | 0 | 8 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ........................................................TTTTTTTTTTCGTTTGTC................................................................................................................................................................................ | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AAGAACTGGCGATGCGGGA.................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAGAATTGGCGATGCGGGA.................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAGAACTGGCGATGCGGGAGG.................................................................................. | 21 | 3 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................ATTGGCGATGCGGGA.................................................................................... | 15 | 1 | 6 | 0.33 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AACTTGCGATGCGGGATGA................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AACTGGCGATGCGGGAGG.................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AATTGGCGATGCGGGATGA................................................................................. | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CTCAGATCAGACGAGAG............................ | 17 | 2 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................CCTCAGATCAGACGAGAG............................ | 18 | 3 | 20 | 0.25 | 5 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................TCAGAACTGGCGATGCGGGA.................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................GAACTTGCGATGCGGGATGAAC............................................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................GACGCGGGAGGAGCAATT........................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................CAGAATTGGCGATGCGGGATG.................................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................ACTTGCGATGCGGGA.................................................................................... | 15 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................AGAACTGGCGATGCGG...................................................................................... | 16 | 2 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................TTGGGATGCGGGAGG.................................................................................. | 15 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................TGCGATGCGGGATGAAC............................................................................... | 17 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGCGTGTTTGTGCGAGAG............................................................................................................................ | 18 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................TCTCAGATCAGACGAG.............................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................GCGTGTTTGGGCGTGA............................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................AGCGTGTTTGTGCGAGA............................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GTCGCGTTGCCTTTCGTTAATATGCTGTATTTTTACAAGAGTGCTGGGTCCATTCGAAAAAAAAAAGCAAACAGAAAAATACGTTCCTGAGCGGAGTTTCCTAAACACTCGCACAAACACGCACACACGGAGCCAGAACTTAATACATTCTTAAACGCTACGCCCTCCTCGTTGACGGGGTCGACCATCGGGTCACTGGATCAGTGAGCCTAGTCTGCTCGCCGGCGGTTCCGCAGGCACATATATATGT
**************************************************....(((((((((..((((...((((..((((((.((((.(((.(((.........)))))).)))).))))))...((....))............))))....))))...)))))))))..(((((......)))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
M024 male body |
GSM343915 embryo |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................TATTCTATATTTTTACTAGAGT................................................................................................................................................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CGGCGGTTCCGCAGGCACATA....... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................GCGGTTCCGCAGGCGCAT........ | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................AAAAAAAAGTAAACAGAAAAACAGGT...................................................................................................................................................................... | 26 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................AAAAAAAAAGGATACAGAAAATT.......................................................................................................................................................................... | 23 | 3 | 10 | 0.20 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| .................................................................................................TTCCTTAACACTCGC.......................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................GGCGCCAGTACTTAAGACA....................................................................................................... | 19 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................TGGGTCCATTCCAAGAA............................................................................................................................................................................................. | 17 | 2 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................TCCGCAGTCAGATATAT.... | 17 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................................................................TCGGGTCACTAGACCAG.............................................. | 17 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................ACAGAGTGCTGGGTCC....................................................................................................................................................................................................... | 16 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................CAGAGTGCTGGGTCGACT.................................................................................................................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..............GGTTAATATGCTGCATATT......................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droSim2 | 3r:22419493-22419742 + | dsi_20682 | CAGCGCAACGGAAAGCAATTATACGACAT---AAAAATGTTCTCACGACCCAGGTAAGCTTTTTTTTTTCG-------TTT-GTCTTTTTA--------TGC---AAGGACTC--GCCTCAA----------------------AGGATTTGTGAGCGTGTTTGTGCGTGTGTGC--------CTCGGTCTTGAATTATGTAAGAATTTGCG--------ATGCGGGAGGAGCAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAGCGGCCGCCAA--GGCGTCCGTGTATAT------------ATACA--- |
| droSec2 | scaffold_13:1732642-1732891 + | CAGCGCAACGGAAAGCAATTATACGACAT---CAAAATGTTCTCACGACCCAGGTAAGCTTTTTTTTATCG-------TTT-AGGTTTTTA--------TGC---AAGGACTC--GCCTCGA----------------------AGGATTTGTGAGCGTGTGT--GT--G----CCTCGGTCTTGAGGGCGCGAATTATGTAAGAATTTGCG--------ATGCGGGAGGAGCAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAGCGGCCGCCAA--GTCGTCCGTGTATAT------------ATACA--- | |
| dm3 | chr3R:22977579-22977828 + | CAGCGCAACGGAAAGCAATTATACGACAT---AAAAATGTTCTCACGACCCAGGTAAGCTTTTCTTTTTCG-------TTT-GGTTTTTTA--------TGC---AAGGACTC--GCCTCGA----------------------AGGATTTGTGTGCGCGTGGGTGTGTGTGTGC--------CTCGGGCGCAAATTATGTAAGATTTCACG--------ATGCGGGAGGAGCAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAGCGGCCGCCAA--GGCGTCCGTGTATAT------------ATACA--- | |
| droEre2 | scaffold_4820:5059669-5059913 - | CAGC-CAACGGAAAGCAATTATACGAAAT---AAAAATGTTCTCACGACCCAGGTAAGCTATATTT--TCG--A-GT----------TTTT--------TGC---AAGGACTC--GCCTCCA----------------------AGGATTTGTTAGTGTGCGT--GT--G-------------TGAGGGCGCGAATTATGTAAGATTTCGCG--------ATGCGGGAGGAGCAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAGCAGCCGCCAA--GGCGTCCGTACATATGTACCTGCATATATACA--- | |
| droYak3 | 3R:23516833-23517068 - | CAAGCCAACGGAAAGCAATTAAACGAAAT---AAAAATGTTCTCACGACCCAGGTAAGCTTTTTTTT-TCGCGAC---TTTCGGT--TTTT--------TGC---AAGGACTC-----TCGA----------------------AGGATTTGTTAGTGTGTGT--G-------------------AAGGCGCGCATTATGTAAGAATTCGCG--------ATGCGGGAGGAGCAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAGCAGCCGCCAA--GGCGTCCGTATATAT------------GTACA--- | |
| droEug1 | scf7180000409770:441524-441753 + | TAACCCAGCGGAAAGCAATTATACGAACC---AAAAATGTTCTCACGACCCAGGTAAGCTTTTT-------------------------TC--------TGT---AAGGACTT--TTCGAGC----------------------AGGATGTGTGAGTGTGAACGTGAATGTT--------TTTTTGGGGCGGGAATTATGCAACATTTCGCG--------ATTCGGGACGAGGAACTTC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ATT------CGGTTCAGACGAGTAGCCGCCAA--GGCGTCGGTATATAT------------AT------ | |
| droBia1 | scf7180000302113:3249938-3250164 + | CAATTTAAAAGAAAGCACT-GTAAAACATAAAAAAGATGTTCTCACGACCCAGGTAAGCTTTTT-----------------------------------CGC---CAGGACTT--TTCGGGC----------------------AGGATGCGCGAGTGTGTGT--GCGTC--------CGCTCTGGGGGCGGGAATTATGCAACATTCGGCG--------ACTCGGGACGAGGAGCTGC-----CCCAGCTGGTAGCCCAGTGACCTAGT-----------------------CGGATCAGACGAATCGCCGCCAA--GGCGACGCTGTGTGT------------GTGCA--- | |
| droTak1 | scf7180000410579:291670-291901 - | TAAGCCAACGGAAAGCAATTCGAGGAATT---AAAAATGTTCTCACGACCCAGGTAAGCTTTTT--------------------------T--------CGC---AAGGACTTTCGGCGAGC----------------------AGGATGTGTGAGTGTGTGG--GT--T---TCCTACGTTTTGGGGGCGGGAATTATGCAACATTTCGCG--------ATTCGGGACGAGGAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ATT------CGGATCAGACGAATCGCCGCCAA--GGCGTCGGTATATAT------------AT------ | |
| droEle1 | scf7180000491280:1023335-1023574 + | TAACCCAACGGAAAGCAATTCTACGAAGT---GAAAATGTTCTCACGACCCAGGTAAGCTTTTTTT-----------------------TT--------TGC---AAGGACTT--CGCGGTC----------------------AGGATGTGTGAGTGAGTAC--GAGTG--T-------TTTCGGGGGCGGGAATTATGCAACATTTCGCG--------ACTCGGGACGAGGAACTGCCCAGCCCCAGCTGGTAGCCCAGTGACCTAGTC-------------ATTCCGTTTCGGATCAGACGAATAGCCGCCAA--GGCGTCGGTATATAT------------AT------ | |
| droRho1 | scf7180000780109:2115-2352 - | GAATCCAACGGAAAGCAATTCTAGGAAGT---AAAAATGTTCTCACGACCCAGGTAAGCTTTTTT------------------------TT--------TGC---AAGGACTT--CGTGGCC----------------------AGGATGTGTGATTGTGTGC--GAGTG--T-------TTTTGGGGGCGGGAATTATGCAACATTTTGCG--------ATTCGGGACGAGGAACTGC-----CCCAGCTGGTAGCCCAGTGACCCAGTC-------------ATTCGTATTCGGATCAGACGAATAGCCGCCAA--GGCGTCGGTATATGT--------------ACATAC | |
| droFic1 | scf7180000453776:152348-152575 + | TTACTTTACGGAAAGCAATAAAACGAATT---AAAAATGTTCTCACGACCCAGGTAAGCTTTT------------------------------------TGC---AAGGACTC--GCGGC-C----------------------AGGATGTG--AGTGTGTGT--GAGTG----C-------TTGGGGGCGGGAATTATGCAACATTTCGCG--------ATTCGGGACGAGGAGCTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ATTCGGATTCGGATCAGACGAGTAGCCGCCAA--GGCGTCGGTATCGGT------------GTACC--- | |
| droKik1 | scf7180000302810:432912-433110 - | CAAC-----GCAAAGCAATTCTAGGAAGG---AACAATGTTGTCACGACCGAGGTAAGCTTTTC---------------------------CT------GGC---CAGG----------CGC----------------------AGGATGTGTGTGCGCGTGT--GT--G-------------TGTTCGCGGTAGTTATGCAATATTTC----------------GGAGGAGGAACTGC-----CCCAGCTGGTAGCCCAGTGACCTAGTC-------------ACT------CGGATCAGACGAATCGCCGCCAA--GGCG-CGGTGTGTGT------------AT------ | |
| droAna3 | scaffold_12911:3895618-3895829 + | TGGCGC-ACGGAAAGCAATTCTAGGAAAC---AAAAATGTTCTCACGACCCAGGTGAGCTTATT----TCG--A-TTT-----GTTTT--CCTTCGCGTGGC---AAGGAGG------ACGA----------------------AGGACGTATGGGTGTGTT----------------------------GTGGATTATGCAACATTGTGA-CAAAGG---AGGAGGAGGAGGAGCTGC-----CCCAGCTGGTAGCCCAGTGACCTAGT----------------------------CAGACGCGAAGCCGCCAA--GACTTCG-TGT------------------------ | |
| droBip1 | scf7180000396640:741279-741490 + | CAACAC-ACGGAAAGCAATTCTAGGAAAT---AAAAATGTTCTCACGACCCAGGTGAGCGTAAT----TCG--A-TTT-----GTTTT--CCTTAGCGGGGC---AAGGAGC------CCGA----------------------AGGACGTTTGGGTGTGCT----------------------------GTGGATTATGCAACATTGTGG-CGGTCGCGGCT---GAGGAGGAGCTGC-----CCCAGCTGGTAGCCCAGTGACCTAGT----------------------------CAGACGCGTAGCCGCCAA--GACTTCG-TGT------------------------ | |
| dp5 | 2:8584111-8584338 + | CATCGAA---AAAAGCAATTCGAGGAAGA---AAAAATGTTTTCACGACCAAGGTAAGCGCTTT--------------------------CCAAGACAATGCAAAAAGGACTC--GACTCCAGTTACAGTTACACGATTTTGGGAGGATT---------------------------------------GGGGCATTATGCAACATGGCGAG--------TGGC------AGGCACTGC-----CCCAGCTGTTAGCCCAGTGACCTAGTCGTTGTCGCGGGGCAGT------TGAGGCAGACGGATGGCCGCCAAAAGACATCTG--------------------------- | |
| droPer2 | scaffold_0:2412436-2412663 + | CATCGAA---AAAAGCAATTTGAGGAAGA---AAAAATGTTTTCACGACCAAGGTAAGCGCTTT--------------------------CCAAGACAATGCAAAAAGGACTC--GACTCCAGTTACAGTTACACGATTTTGGGAGGATT---------------------------------------GGGGCATTATGCAACATGGCGAG--------TGGC------AGGCACTGC-----CCCAGCTGTTAGCCCAGTGACCTAGTCGATGTCGCGGGGCAGT------CGAGGCAGACGGATGGCCGCCAAAAGACATCTG--------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 03:25 PM