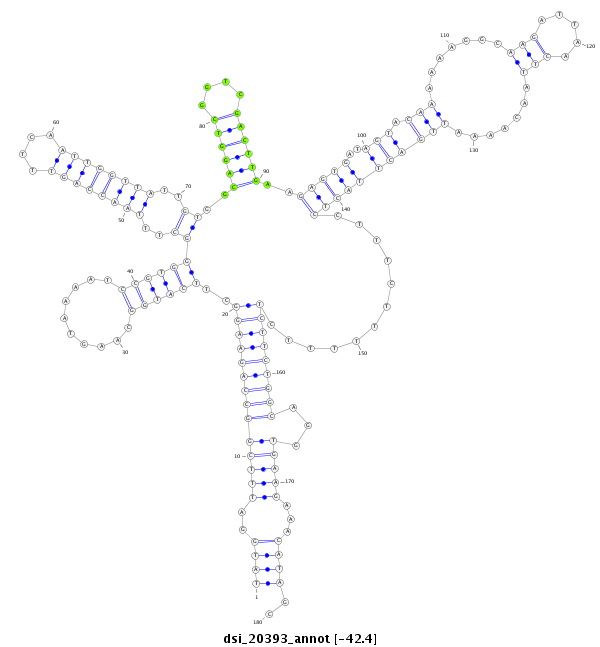
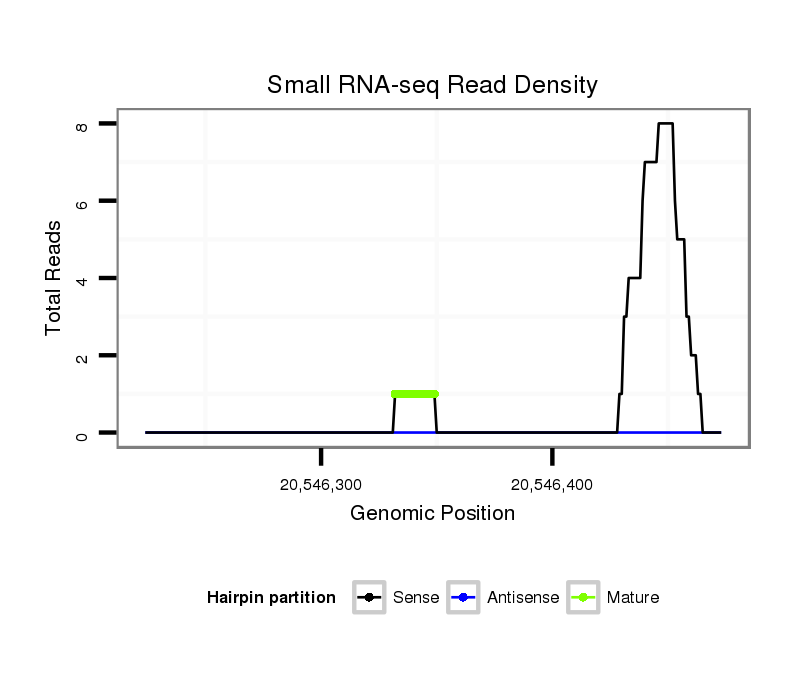
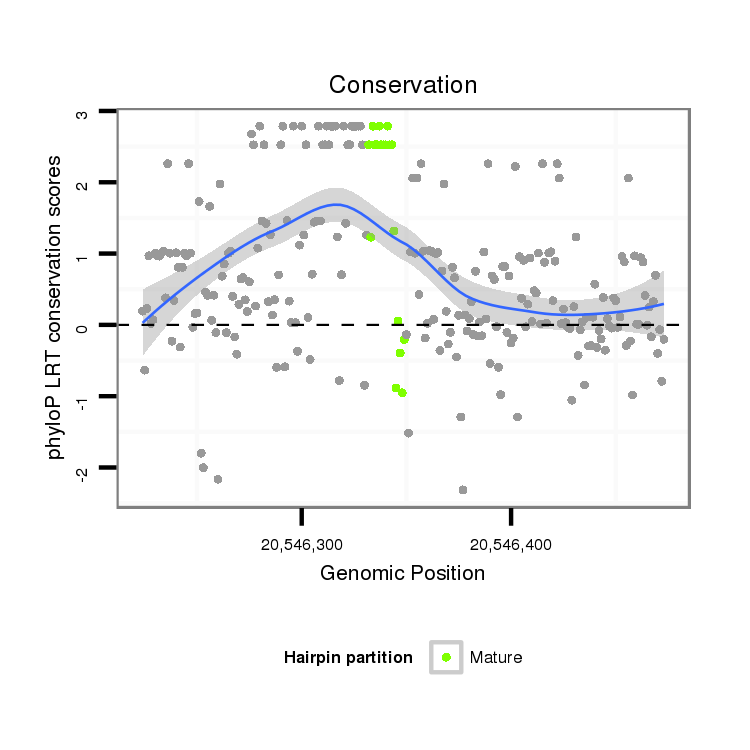
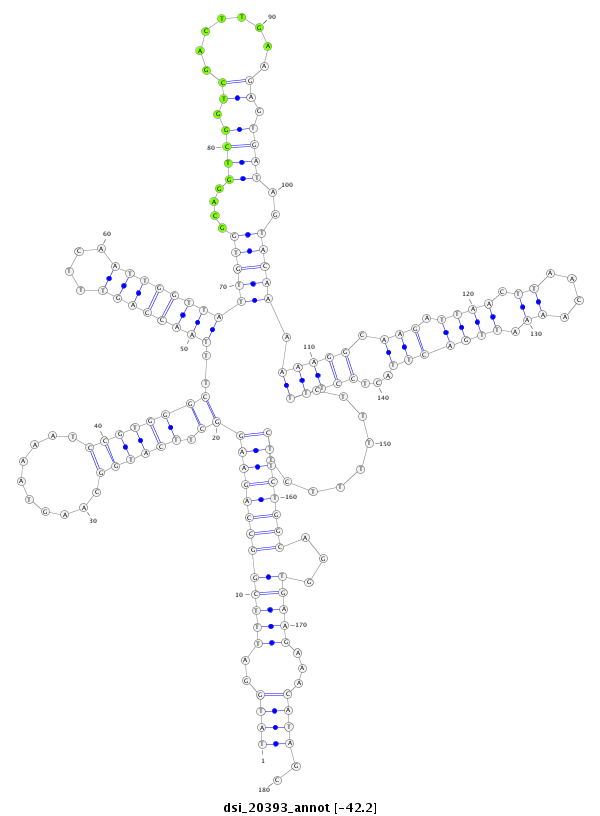
| AAGTCGATCGATTAAAGTGTGAATTATGG----------CAT------------------GGATTTTCGACTGAGTAAACACCAATAGATCTACAGATGATCTGTATGATGATGC-----ACCCAGTCTTCTTGGTTAGTGAAATGTGTGGGTTTAATCCAGTTTCGATTGGTTATTGTGGAAGGTCGGTCGACCGGCGTGTG---TAAGGGAGTGATAACAAAA-AT----TAAGA-A-----AAAAT------------CCAAAAAA----ATAATTCAATA-----------TTTGTTA--ACTC--TTTCCTTGCAGAAA--------------------------------------------------------------------CTAAGCGAACAAGA-----AGGAGGCTTTAAGGAGTAACCAAGAACCTATGCCTATGCCTATGGAGTAGCCCAATTGAGC-----CACGAAG-- | Size | Hit Count | Total Norm | Total | M022
Male-body | M040
Female-body | M059
Embryo | M062
Head | V043
Embryo | V051
Head | V112
Male-body | SRR902010
Ovary | SRR902011
Testis | SRR902012
CNS imaginal disc |
| ..........................................................................................................................................................................................................................................................................................TA-----------TTTGTTA--ACTC--TTTCCTTGCA...................................................................................................................................................................... | 23 | 1 | 3.00 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTTGTTA--ACTC--TTTCCTTGCAGT.................................................................................................................................................................... | 23 | 1 | 3.00 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTTGTTA--ACTC--TTTCCTTGCAG..................................................................................................................................................................... | 22 | 1 | 3.00 | 3 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................TA-----------TTTGTTA--ACTC--TTTCCTTGCAG..................................................................................................................................................................... | 24 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................TA-----------TTTGTTA--ACTC--TTTCCTTGC....................................................................................................................................................................... | 22 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................AGAAA--------------------------------------------------------------------CTAAGCGAACAAGA-----AGGAGG..................................................................... | 25 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTTGTTA--ACTC--TTTCCTTGCA...................................................................................................................................................................... | 21 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................TA-----------TTTGTTA--ACTC--TTTCCTTG........................................................................................................................................................................ | 21 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CTTTAAGGAGTAACCAAGAACCT.............................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................AGCGAACAAGA-----AGGAGGCTTTA................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................AACCAAGAACCTATGCCTATGCCT.................................. | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTATGGAGTAGCCCAATTGAGC-----......... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................AA--------------------------------------------------------------------CTAAGCGAACAAGA-----AGGAGGC.................................................................... | 23 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................TTTGTTA--ACTC--TTTCCTTGC....................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................GGAGGCTTTAAGGAGTAACCAAGAACC............................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................GAAA--------------------------------------------------------------------CTAAGCGAACAAGA-----AGGAGG..................................................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................GAGTAGCCCAATTGAGC-----C........ | 18 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................GTAACCAAGAACCTATGCCTATGC.................................... | 24 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................GA-----AGGAGGCTTTAAGGAGTAACC...................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................GA-----AGGAGGCTTTAAGGAGT.......................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................AAACACCAATAGATCTACAGATG................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .AGTCGATCGATTAAAGTGTGAAT.............................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................GGAGTAACCAAGAACCTATGCCT........................................ | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................AAGCGAACAAGA-----AGGAGGC.................................................................... | 19 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................ATGCCTATGGAGTAGCCCAATTGAGC-----......... | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................TA-----------TTTGTTA--ACTC--TTTCCTGC........................................................................................................................................................................ | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CTTTAAGGAGTAACCAAGAACCTATGC.......................................... | 27 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |