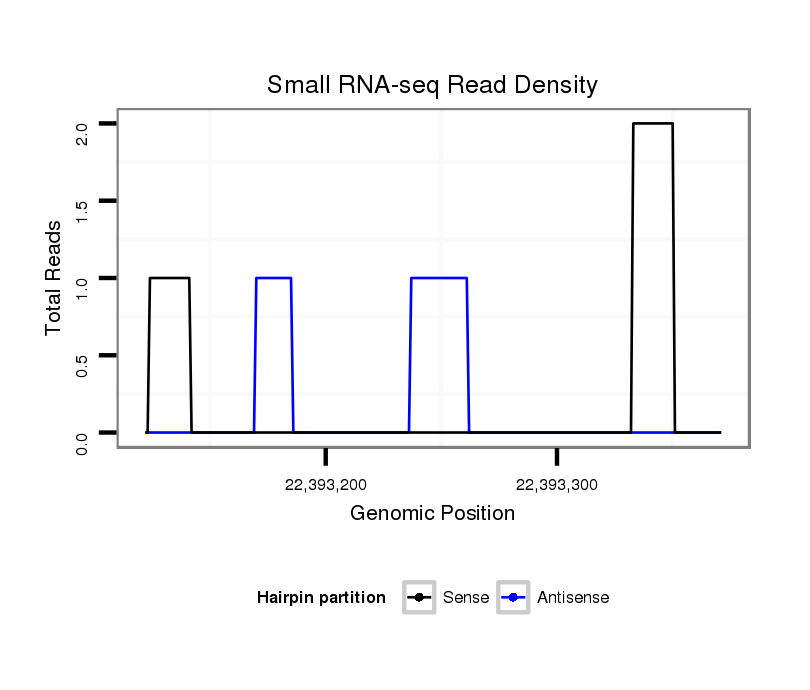
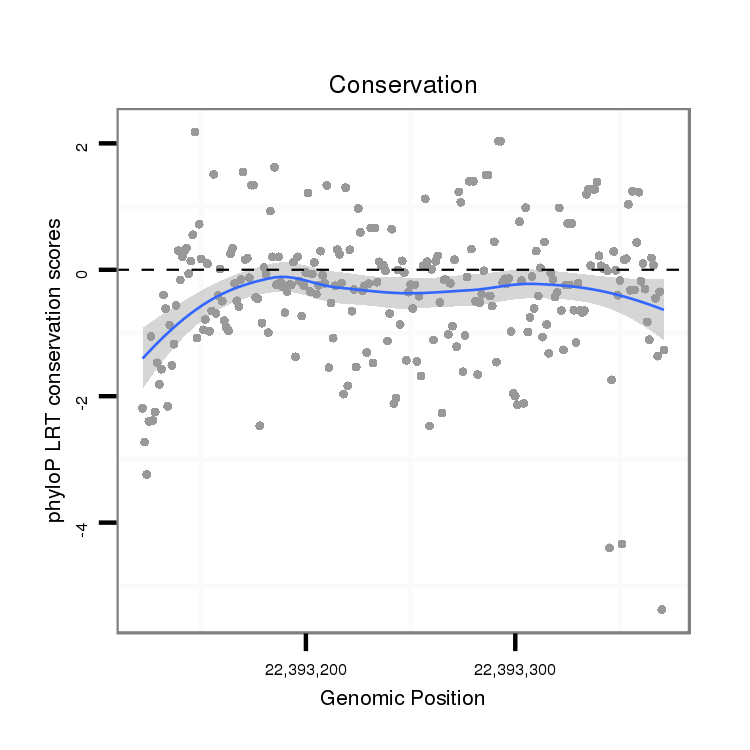
| droSim2 |
3l:22393122-22393371 + |
dsi_20377 |
GAGTACTA-CGATGCCATGGCCTT--CCTCAAGGAGGTCATTCGAGAACGCAAG----TAAGAC-GACGTTTGACCTGCCCCAAG------------ATACTAGCTTATGA--------GATAGA----TACCGTTTAAGGTTAGAAGA-------------------------GCTCAGAGT-ATAGCT-------AGGTTGCATT------------------------------------------------------------------TCTCTAATGTC------------------------------------------------------------AA-------------------------TTTTGGACATT-CCTGTTATTT------------GGCAAAGTC--GTGTTTGTGGTTTTTGGAACATTTCGC---------------------------------------------------------------GATATTT-------------------------------------------------------------------------------------------------------------------------------ATTTCTTTTTATTTC---------------TGTACCC---------GTTACTATT |
| droSec2 |
scaffold_11:2807338-2807615 + |
|
GAATACTA-CGATGCCATGGCCTT--CCTCAAGGAGGTCATTCGAGAACGCAAG----TAAGAT-GACGTTTGACCTGCCCCAAG------------ATACTAGCTTATGA--------GATAGG----TACCGATTAAGGTTAAAAGA-------------------------GTTGAGGGT-ATAGCG-------AGGTTTCATC------------------------------------------------------------------TCTCCAATGTC------------------------------------------------------------AA-------------------------TTTTGGACATT-CCTGTTATTT------------GGCAAAGTC--GTGTTTGTGGTTTTTGCAACATTTCACGATACGAAAGATAATTAAGCTTTAGTTAGACCATACGGGC---------------------------------------------------------------------------------------------------------------------------TG------------------------------------GATTTTATTTC---------------TATACTC---------GTTA---CT |
| dm3 |
chr3L:22905284-22905436 + |
|
GAATACTA-CGATGCCATGGCCTT--CCTCAAGGAGGTCATTCGGGAACGCAAG----TAAGAC-GACGTTTGACCTGCCCTAAA------------ATACTACCTTATGA--------TATATGTAGATCCCGTTTAAGG-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GA-------------------------TTTTGGACATT-CCAGTTATTT------------GT-----------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTAGTGTCTT---------------TG---------------------GA |
| droEre2 |
scaffold_4784:22554468-22554661 + |
|
GAATACTA-CGATGCCATGGCCTT--CCTCAAGGAGGTCATTCGGGAGCGCAAG----TAAGAC-GATGTTTGACCTGCCCTAAG------------CTATTAGCTTATCA--------GA-------------------------AGT-------------------------GCTTGGGGT-TCAACT--------GGTAACCTT------------------------------------------------------------------CCTCCAATGTC------------------------------------------------------------CA-------------------------TTTTGCACATT-CCTGTTATTC------------GACGAAATC--GTGTTTGTGTTTTATACTACACGTTGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATA |
| droYak3 |
v2_chrUn_3594:560-767 + |
|
GAATACTA-CGATGCCATGGCCTT--CCTCAAGGAGGTCATTCGGGAGCGCAAG----TAAGAA-GATATTTGACCTGCCCCAAG------------CTATTAGCTTATGA--------GATA-A----TCCTGCTTAAAGTCAGA--T-------------------------GCTTAGGGT-ATAATT------------ACCTT------------------------------------------------------------------TTTCCAATGTC------------------------------------------------------------CA-------------------------TTTTGGACATT-TCTTTTATTC------------GATTAATTC--GAGTTTGTGGTTTATACAACACGTTGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATA |
| droEug1 |
scf7180000409589:16724-16860 - |
|
AGCTCGAA-GCTTTT-G---------------------------------------------------------------------------------------TTGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAAGCTGAATTTCTAGCATTTTCGGCGTGGTT----------------------TTGTT--------GCGAAGTT-----------------------------------------GATTTGGTTTTGATTTGAAC--TCGGAATAA-----------AAT---TCGGAAATTT-------------------------------------------------------------------------------------------------------------------------------ACCTGCCCCGGTTTT---------------TATACCC---------GTTACTCGT |
| droBia1 |
scf7180000302514:12131-12283 + |
|
TGTGAGGG-GAATTCGG---------------------------------------------------------------------------------------------------------------------TTTCTATTTAGATGA-------------------------GTTTGG-----------------------------------------------------------------------------------------------------------------------------------------------------------C-TATATGTTGAATTATTATTATTTTTGTAATGTTA----------------------TTGTTAATAATA-------TTTTT----TATTGTTGTT---------------------------------------GTAAC--TTTGATTAC--------------------------------------------------------------------------------------------------------------------------G------------------------------------ATTTTTTTTG-----TGTATTT---TCTGAATTCCTTC---GACTATAGT |
| droTak1 |
scf7180000414699:7546-7811 - |
|
GCGGAACA-CAATCTCTGGACCTT--TCTGAGCCATTTTGTCTAAGAAACTATGCCTTGAAAAT-CACGTTTGACCCTCCGCAATTGCAAAAGAAAAATACTGGTTTTTCG--------TTTAT---------------TTTGAGAAAA-------------------------AT-ATCA----------TAGTAG----------CCGTTACAGATATTTATCAAATTTAA----------------TCTCTCTTTTTTATTAGAAAATTTGCT-------------------------------------------------CTCCCTTTAGTTTATTTCTTCAAAT-------------------------ATGTTTT-TTTATTTCAT------------AC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TGTTTTTT---TATTATTTCCCAGACC-----CAAATA---AGACC |
| droEle1 |
scf7180000488055:5364-5530 - |
|
ACGCAAG---------------------------------------------------TAAATCATACTTTTAATGTTTTC-AGG------------ATATT-------------------TAGT----TT------------------TTGCAATAGCTGCAAGGGTATATGAACTTCGG-C-TTGGCG-------AAGTTT-GCT------------------------------------------------------------------TCCTTAATTT--------------------------------------------------------------T-------------------------ATTTATATATT-TATTTTATTT------------AATAATATTTATTATTTGTAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATTTCTTTTACTTTT---------------TATACC------------------C |
| droRho1 |
scf7180000768839:20972-21157 + |
|
GAAATAGCACCGT---------------TTAATAAAATTATAAT-----------------------------------------------------------------------------------------------------------------------------------------GT-A-----AAG-AGG----------TATACGTGTT-------------------TCTTTA---------------ACATTTAAGACATGTTTGCTTGTTCTCTCTGATTCCATTTTGTAAAGTAATGTAAA-----------------------------------------------------------------------------------------------------------ATGTAAAT-----------------------------------------------------------------------------------TTAAATACATATTTTGTTTTATCCGTTTTTAT-----------------------------------------------------------------T------------------------------------TTTTTGGT---CATGTAATTTT---TATACCC---------GTTACTCGT |
| droFic1 |
scf7180000453830:115903-115987 - |
|
GAGTACTA-TGACGCTATGGCCTT--TCTAAAGGAAGTCATTCGGGAACGCAAG----TAAGGAGGGCTTCTAGCCAGTTCAGAG------------ACACAAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000302384:217458-217611 + |
|
AAATACCC-TGCCA---------GGGTATAAAAATTATAGC------------------------------T-----------------------------------------------------------------------------------------------------------------------------TCGTT-----------------------------------------------------GT-TTTTCAACGTATGTTTATCTACTCTC------------------------------------------------------------CG-------------------------ATATAAGCATTTTTTATTATTC------------TAGAAATTC--GTATTAATTTTAT---GAAAATCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GG---CGACTATATCT---TATAGCTGCCATAGGAA---CGATC |
| droAna3 |
scaffold_13417:5942538-5942735 + |
|
ACGACGT---------------------------------------------------------------TTGATTCGCCCCATA------------TCACAATCCTATGC--------GATAGG----TA------------------TTGTGATTCCAGCATGGCTAATTGAAGTTTGG-CGGTGGC-------------------------GGATCTGTAATGGCCTTCA------------------------------------------------------GCCTTCATTAAGTTCCAAATACTGACATATAATTTTGAGTGAACTTGATTCATTTTTCC-------------------------ATTTCCATATT-TTTTTTATTT------------AA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTT---------------TT-----------------TATTAT |
| droBip1 |
scf7180000394386:21990-22160 - |
|
TATCAGTTATTGT---------------TAAAGGA-------------------------------------------------------------------------------------------------------------------------------------------GTTT------------------------TTATTTTTTCTTGT--------CTGATTTTTTGATTTTTG---------------TCATTTTGGACATTTTTGT-------------------------------------------------ATGTATTCAGT-AATATCTTGAGACAT----ACTTTT-------------------------------CCAATAATAATG-------TCTTT------------------------------------------------------TGAC--CTTGGCCAA--------------------------------------------------------------------------------------------------------------------------G-----------GTTTTTGAGATGCTTGTGTG---------------------------------ACCT---------CCTTGTATT |
| dp5 |
XR_group5:130483-130603 - |
|
AGTATGGC---------------------------------------------------------------T-----------------------------------------------------------------------------------------------------------------------------GAGTT-----------------------------------------------------TT-TC-------------TC----------------------------------------------------------------------------------------------------------------------CCACTAATAGTA-------TTTTT----AGCTTCGGTT---------------------------------------GCGTT--GCCGGCCGG--------------------------------------------------------------------------------------------------------------------------TACTTTTGCAGAATTATCAAAATGTTTTGTTGGTTTCTTGGTTTTTT---------------TATACCC---------GATA---CT |
| droPer2 |
scaffold_51:48987-49156 + |
|
CCAAGTTA-CGAACTAGCGGACTG--TATTAT-----------------------------------------------------------------------------------------------------CTT-----------------------------------------TAATAT-TGATTT--------TATTGTATT------------------------------------------------------------------CAGCCAGGATC------------------------------------------------------------CG-------------------------GTTTTCTCTTT-CCTATTTACT------------TTTGAAATC--ACATAAA-----------------------------------------------------------------------------------------------------TTGGTGCCCCCGTTTTCACATTCCGCAAGATTTCT-----------------------------------------------------------------------------------TCGTTTTCACGCC----------------AAAAA-----CATATATTACCATT |
| droWil2 |
scf2_1100000004958:40325-40502 + |
|
ACATGTTC-CAAAAACCAATCTATGACATAGAAATGGTCTTTCTTTGTTTCAAA----AAA-----------------------------------------------------------------------------------------------------------------GCTGA------------TAATAT----------CTTCTACACAGACGTAGCA----------------GAATTTCCC----------------------TACTTG------------------------------------------------------------------------------------------------------------TTCTCT------------GAGCACGTC---------------------------------------------------------------GTCGATTGCTCTTTGGAAAACAC---TTTGAAATTT------------------------------------------------------------------------------------------------------------------------TCCGT-------TGTTCATTTT---------------TTTATCA---------ACA--ACTC |
| droVir3 |
scaffold_13050:2210900-2211033 - |
|
ATACTGGCGAATGCTGG--------------------------------------------------------------------------------------------------------------------------------TAGA-------------------------TCGCAGTGT-A-----TTG-CAT----------TTACCGCGTCTATGTTGAT----------------GAGTGGCCT----------------------GATTGT------------------------------------------------------------------------------------------------------------TTTGAA------------TAGGGTGTT---------------------------------------------------------------TTTGTTTGTGAGTTTTATA-------------------------------------------------------AGGTGGCG-----------------------------------------------TA------------------------------------TTTTTTATTTG---------------TATACC-----------------CT |
| droMoj3 |
scaffold_6680:23213506-23213637 - |
|
TTAAAAGA-GCT---AC-------AATATG----------GCGATGAAAAAA--------------------------------------------------------------------------------------------------------------------------ACTA------------TTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------CAATTTTCGC-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGCCACTCAGTGATTTCAATTTTTTATTAGAAAAAGTACATTTTTTCAGCAATTTCGCGATAAG------------------------------------CATTTTGTTTT---------------TATACCC-----------------T |
| droGri2 |
scaffold_15245:6020970-6021109 + |
|
TCAGAGAA-TATCTCGG---------------------------------------------------------------------------------------TTATTCATTTGGTTTTGCCGG----TG------------------CTCTGACATCTC----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TC----------------------TTGTT-------GGCAAAAGG--TTGGCTCTGGTT------------------------------------------TTAGC--CCAGATTAC-----------GTTTGCCAAGAAATTC------------------------------------------------------------------------------------------------------------------------TTTGT------TTTTCTCTTTT---------------TATACCC---------ATTA-AATA |