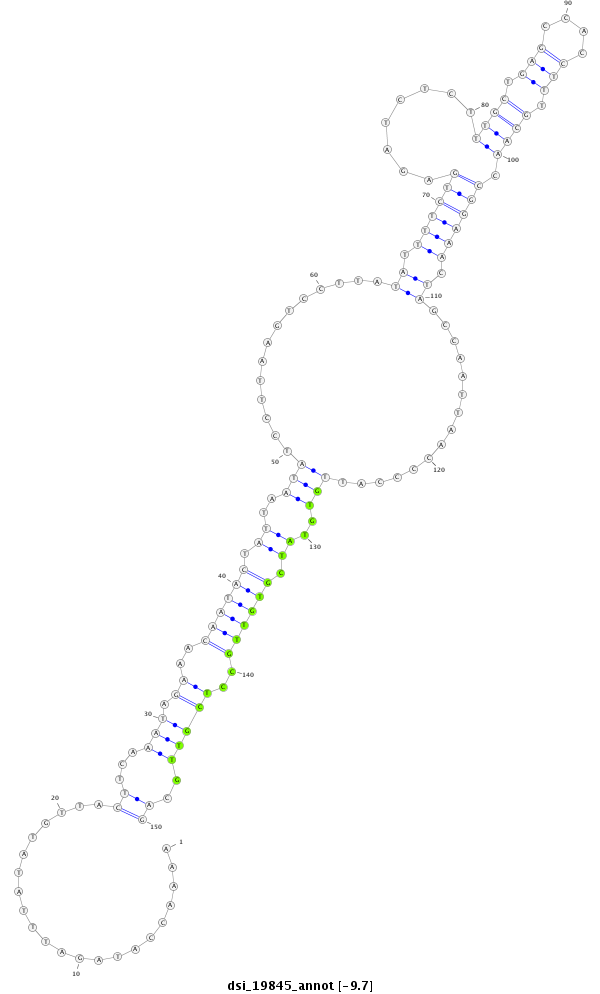
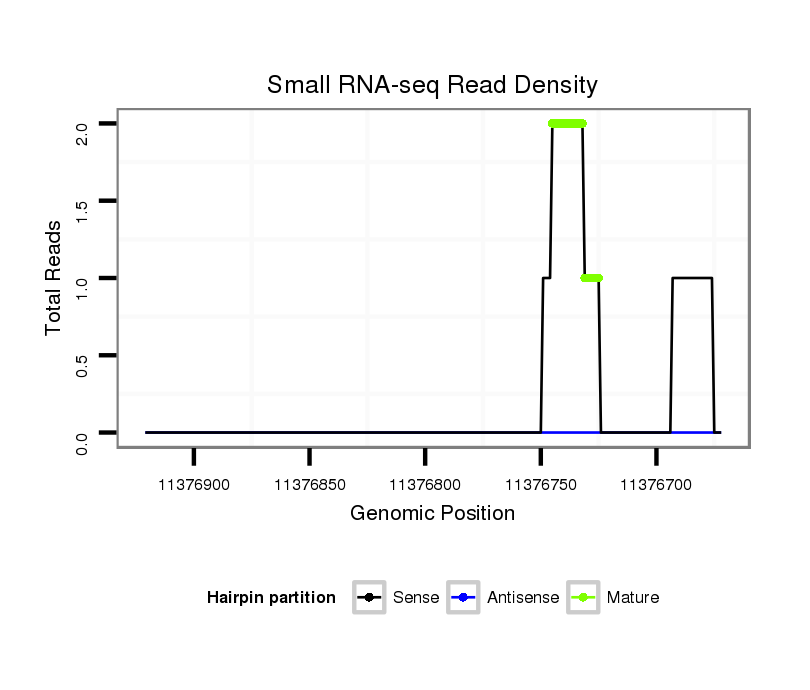
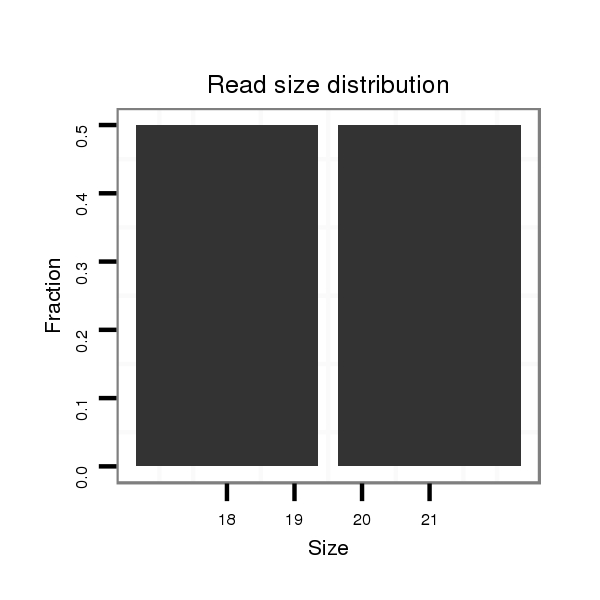
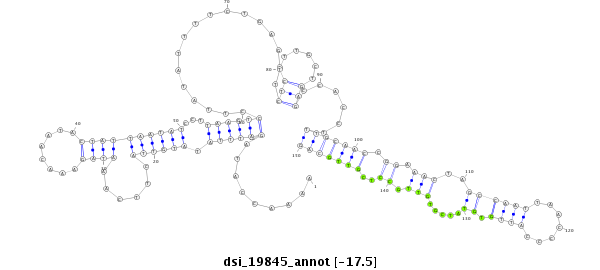
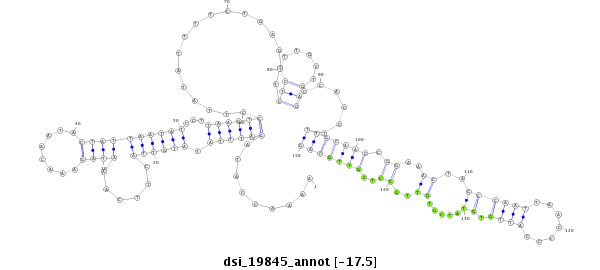
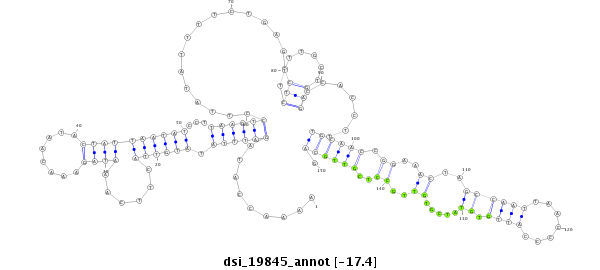
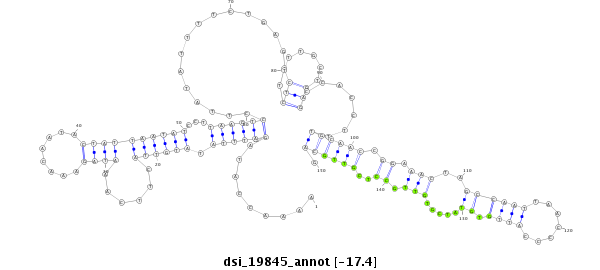
ID:dsi_19845 |
Coordinate:3l:11376722-11376871 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -17.5 | -17.5 | -17.4 | -17.4 |
 |
 |
 |
 |
five_prime_UTR [3l_11376538_11376721_-]; exon [3l_11375323_11376721_-]; intron [3l_11376722_11380942_-]
No Repeatable elements found
| --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TCTTAAAAAGCTAACAAGTAGAAGACCAAAAAATACCCTTTACTTTACAGAAAACCATAGATTTATATGTTACTTCAAATAGAAACAATACTATTAATATCCTTAAGTCCTTATATTTTCTGAGATCTCTTTGCTGAGCCACCTTTGCAACCGGAAACTAGCCAATTAACCCCATTGTGTATCGTGTTGCCTCGTTGCAGGTGAAACTCGCTAATTGTCGGGTGGAAAAAGGTTACCAATACACAGGGCA **************************************************......................((...(((.((..((((((.((..(((..............((.((((((........((((.(((....))).)))).)))))).))...............)))..)).))))))..)))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M025 embryo |
O001 Testis |
O002 Head |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
SRR553486 Makindu_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................GTGTATCGTGTTGCCTCGTTG..................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................AAGGTTACCAATACACAG.... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................TGAGATCGCTTTGATGAGCTA............................................................................................................. | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................CATTGTGTATCGTGTTGC............................................................ | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TGGCGGGTGGAAAAAGGTGA............... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TGAGATCACTTTGATGAGCTA............................................................................................................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................CTGAGAGCTCTTTGTTGA................................................................................................................. | 18 | 2 | 11 | 0.18 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| ...................................................................................................................................................................................................................TAAGGGTCGGGTAGAAAAA.................... | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................GTGGAGAAGGGTTACCA............ | 17 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................CTCGCTGATTGTAGTGTGG......................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................TAAGTTCTTATATTTTTTGA............................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AGAATTTTTCGATTGTTCATCTTCTGGTTTTTTATGGGAAATGAAATGTCTTTTGGTATCTAAATATACAATGAAGTTTATCTTTGTTATGATAATTATAGGAATTCAGGAATATAAAAGACTCTAGAGAAACGACTCGGTGGAAACGTTGGCCTTTGATCGGTTAATTGGGGTAACACATAGCACAACGGAGCAACGTCCACTTTGAGCGATTAACAGCCCACCTTTTTCCAATGGTTATGTGTCCCGT
**************************************************......................((...(((.((..((((((.((..(((..............((.((((((........((((.(((....))).)))).)))))).))...............)))..)).))))))..)))))..))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR902009 testis |
M024 male body |
SRR553485 Chicharo_3 day-old ovaries |
SRR553486 Makindu_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................TTCTGGTTTTTTATGGAAA.................................................................................................................................................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........CGTTTGTTCATCCTCAGGTTT............................................................................................................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................AGGAATATATAAGACTCAA............................................................................................................................ | 19 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................TCTTTGTTAGGATAGTTAT....................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........GATTGTTCCTCGTCTGGTT............................................................................................................................................................................................................................. | 19 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................CCTTTGATCGGGTAGATGG............................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................ATATAAAGGACTCTAGAGA........................................................................................................................ | 19 | 1 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................GGAGCTACGTCCACCTT............................................ | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/24/2015 at 02:47 AM