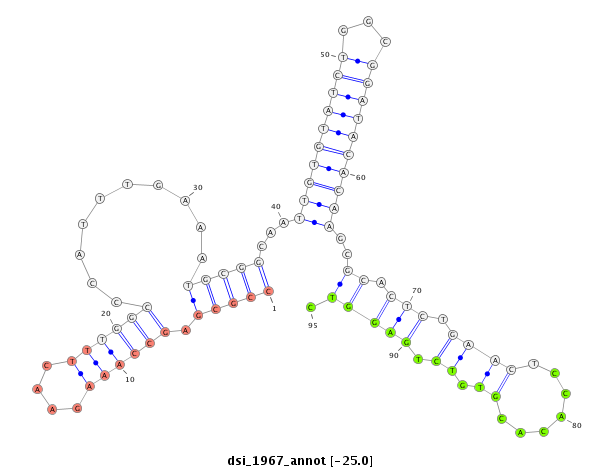
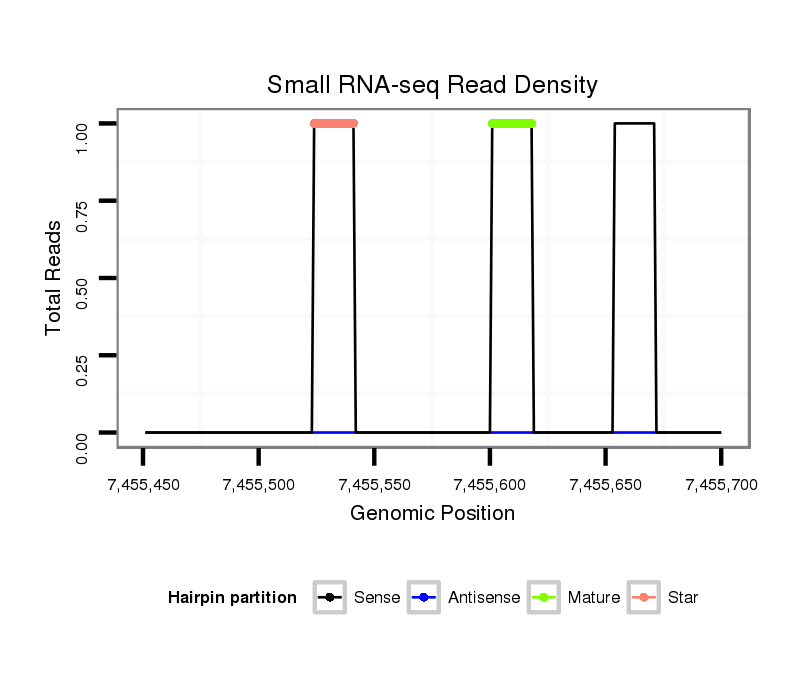
ID:dsi_1967 |
Coordinate:3l:7455501-7455650 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
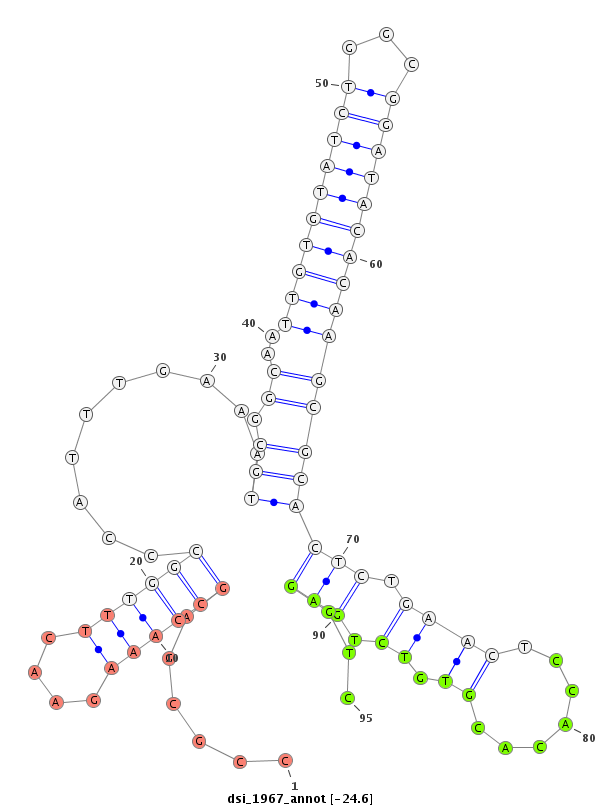
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -24.6 | -24.4 | -24.3 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCTGTTGCCAGTGGTAACCGAAGCAATTGTGTTTAATGCGGCTCTTTGCATTGGCCCATTGCCTGTGCATGTGCCGCGAGCCAAAGAACTTTGGCCCATTTGAAATGCGGCAATTGTGTATCTGGCGGATACACAAGCGCACTCTGAACTCCACACGTGTCTGAGGTCTTTTTCTTAGCTTTTTTCCTCTGCATTTTCAGGCTGACACGAACATCGAGACAGGGCGAGCAGAAGCAGCTGTTACTGGTTC
*************************************************************************(((((.((((((....))))))..........)))))...((((((((((...))))))))))..((.(((.((((.......)).)).))))).********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
O001 Testis |
O002 Head |
SRR553488 RT_0-2 hours eggs |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
M024 male body |
SRR902009 testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................CCACACGTGTCTGAGGTC.................................................................................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................GACACGAACATCGAGACA............................. | 18 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................CCGCGAGCCAAAGAACTT............................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................TGAGATCTTTTTGTTAGCTGTTT.................................................................. | 23 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTTTCGGAGGGCTTTTTCTT.......................................................................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GAAGAACGTTGACCCATTTGA................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................CGTTTCGGAGGGCTTTTTCT........................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAGCCCATTTGAAAT................................................................................................................................................ | 15 | 1 | 20 | 0.15 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................AGACAGGTCGAGCAGGAAC............... | 19 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................AAGTAGCTGACACTGGTTC | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................CGCGTCTGCGGGCTTTTTCT........................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CTTAGCTTATTTCCT.............................................................. | 15 | 1 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................GCTACGCAAGAGCACTCT......................................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................TTTCAGGCTGATGCGAA....................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
|
GGACAACGGTCACCATTGGCTTCGTTAACACAAATTACGCCGAGAAACGTAACCGGGTAACGGACACGTACACGGCGCTCGGTTTCTTGAAACCGGGTAAACTTTACGCCGTTAACACATAGACCGCCTATGTGTTCGCGTGAGACTTGAGGTGTGCACAGACTCCAGAAAAAGAATCGAAAAAAGGAGACGTAAAAGTCCGACTGTGCTTGTAGCTCTGTCCCGCTCGTCTTCGTCGACAATGACCAAG
**********************************************************************************(((((.((((((....))))))..........)))))...((((((((((...))))))))))..((.(((.((((.......)).)).))))).************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................TGCACAGACTCCAGAG................................................................................ | 16 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| .......................................................................................................................................TCGCGTGAGACTGGAGG.................................................................................................. | 17 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ............................................................................................................................................................................................................CATGCTTGTAGCTCTATCCC.......................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 |
| ...................................................................................................................................................................................AAAAAAGTAGATGTAGAAGTC.................................................. | 21 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 |
Generated: 04/24/2015 at 09:41 AM