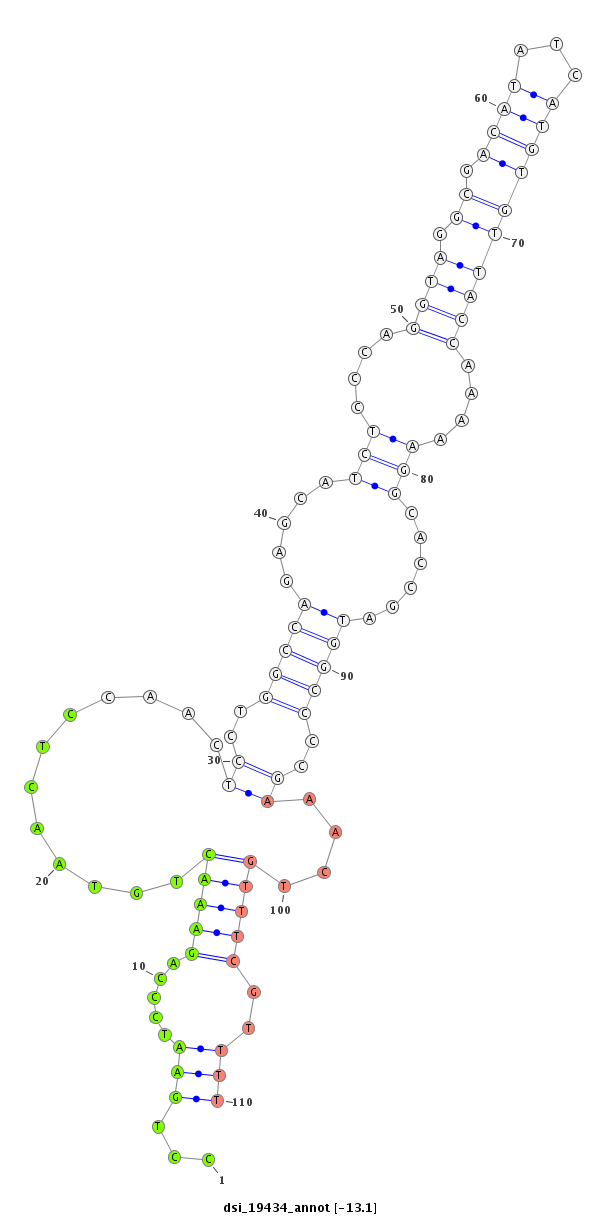



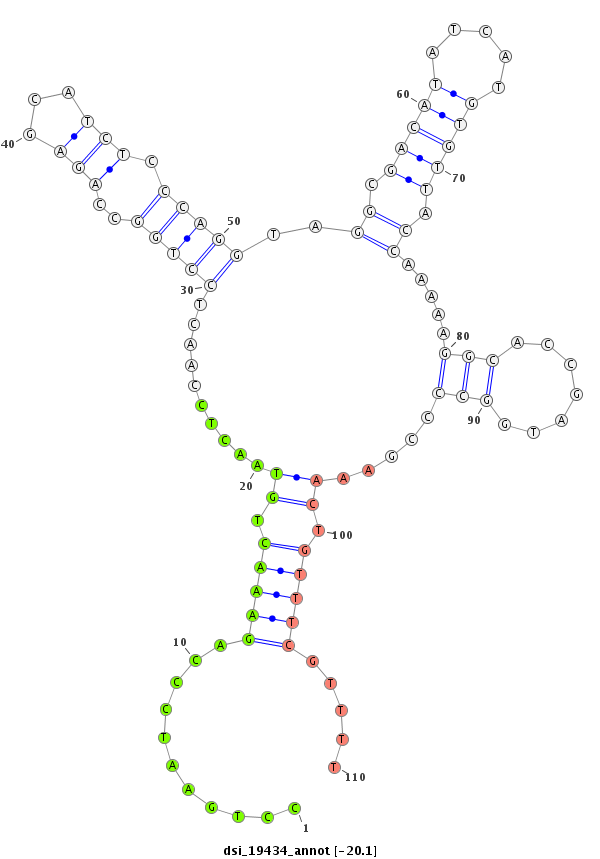
ID:dsi_19434 |
Coordinate:3r:17539804-17539953 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
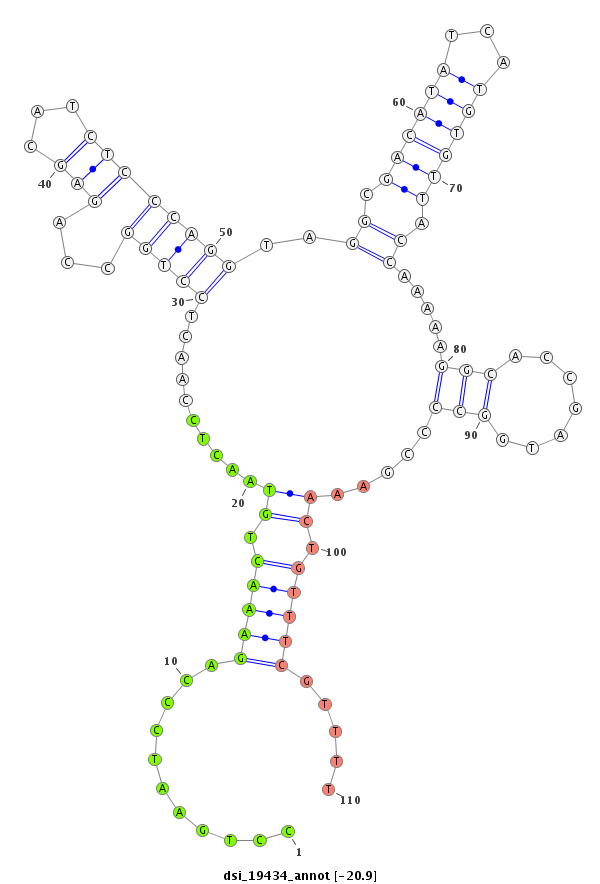
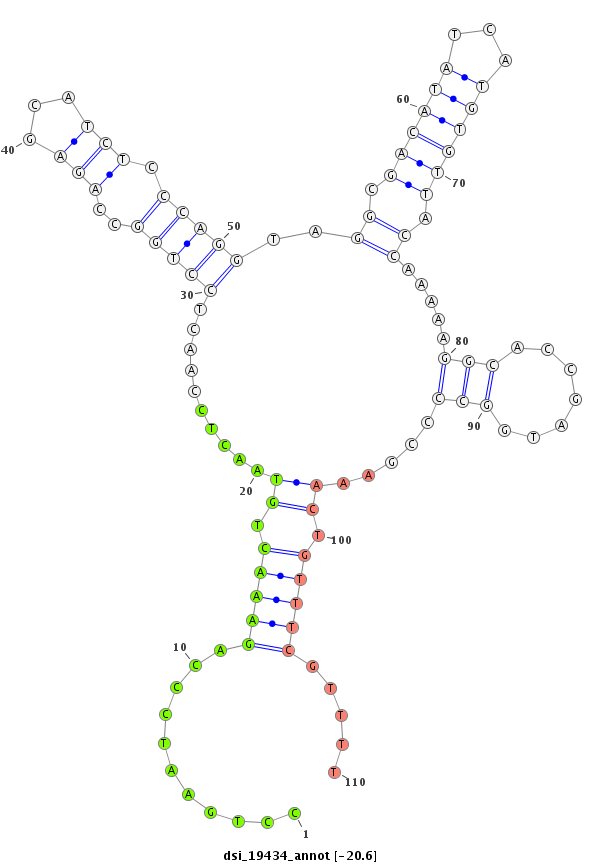
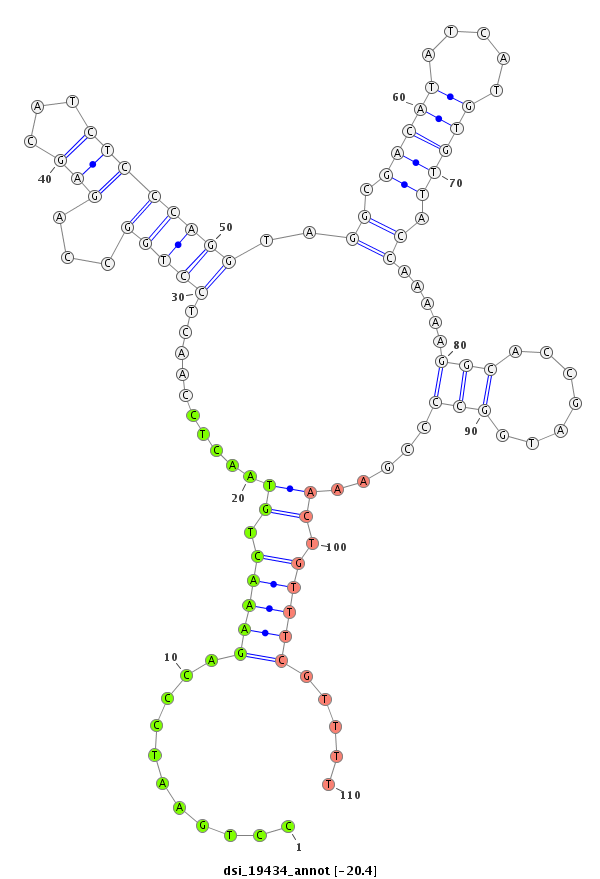
| -20.9 | -20.6 | -20.4 | -20.1 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
TCCATGCCGGTCTCCATTGTTCTCCACTCAGTGAGTGCTTCTCTCTGCCTGCTTCCCTTCTCGTCGTCATTGTACCCGAATCCCTGAATCCCAGAAACTGTAACTCCAACTCCTGGCCAGAGCATCTCCCAGGTAGGCGACATATCATGTGTTACCAAAAAGGCACCGATGGCCCCGAAACTGTTTCGTTTTCTGCCCAGCAAATCCACAAGGTGCACTGAGAGTAAAGTGTTGCAACAAAAAATGTTAA
**********************************************************************************...(((.....(((((............((..(((((.....(((....((((.((.((((...))))))))))....)))......)))))..))....)))))..)))********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR553485 Chicharo_3 day-old ovaries |
SRR618934 dsim w501 ovaries |
GSM343915 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................CCTGAATCCCAGAAACTGTAACTC................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................................AGAGCATCTCCAAGGT.................................................................................................................... | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................AAACTGTTTCGTTTT.......................................................... | 15 | 0 | 5 | 0.40 | 2 | 0 | 0 | 2 | 0 |
| ...........................................TCTGCCGCCTTCCCTCCTCGT.......................................................................................................................................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................GCCCGAGCATATCCCAGCT.................................................................................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 |
| ........................................................................................TCCCAGAAACTGAAA................................................................................................................................................... | 15 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................GATTTCTGCCCAGCA................................................ | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
AGGTACGGCCAGAGGTAACAAGAGGTGAGTCACTCACGAAGAGAGACGGACGAAGGGAAGAGCAGCAGTAACATGGGCTTAGGGACTTAGGGTCTTTGACATTGAGGTTGAGGACCGGTCTCGTAGAGGGTCCATCCGCTGTATAGTACACAATGGTTTTTCCGTGGCTACCGGGGCTTTGACAAAGCAAAAGACGGGTCGTTTAGGTGTTCCACGTGACTCTCATTTCACAACGTTGTTTTTTACAATT
**********************************************************...(((.....(((((............((..(((((.....(((....((((.((.((((...))))))))))....)))......)))))..))....)))))..)))********************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
M024 male body |
SRR553488 RT_0-2 hours eggs |
SRR553487 NRT_0-2 hours eggs |
SRR902009 testis |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................AAGAGAGACGGACGAA.................................................................................................................................................................................................... | 16 | 0 | 3 | 3.00 | 9 | 9 | 0 | 0 | 0 | 0 |
| ..............................................................................TTAGGGACTTAGGGT............................................................................................................................................................. | 15 | 0 | 3 | 1.00 | 3 | 2 | 0 | 1 | 0 | 0 |
| ................................................................................................................................GGTCCAGCCGATGCATAGTA...................................................................................................... | 20 | 3 | 3 | 1.00 | 3 | 0 | 3 | 0 | 0 | 0 |
| .................................................................................................................................GTCCATCCGATGCATAGTAA..................................................................................................... | 20 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................GGTCCATCCGATGCATAG........................................................................................................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................GGATTTAGGATCTTTGACAAT................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................AAGAGAGACGGACGAAGGAG................................................................................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TCCATCCGATGCATAGTAG..................................................................................................... | 19 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................TCCATCCGATGCATAGTAA..................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................GGTCCAGCCGATGCATAGT....................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................GGGATGAGCAGAAGTAA................................................................................................................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................GTAGGGAACAGCAGCAG...................................................................................................................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................GGATGAGCAGTAGTAAC.................................................................................................................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................AGGGTCTTTGACCTCGAAGT.............................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................CATAAGGACGAAGGGAAGA............................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
Generated: 04/23/2015 at 11:08 PM