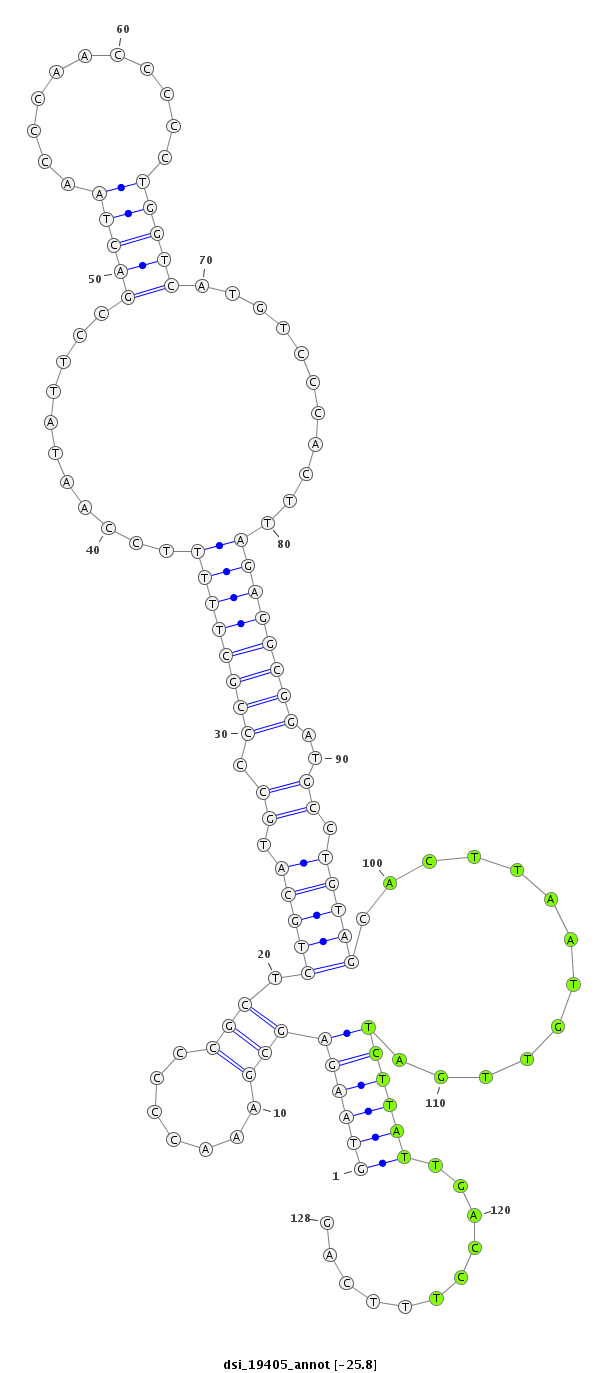



ID:dsi_19405 |
Coordinate:2r:4783798-4783925 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
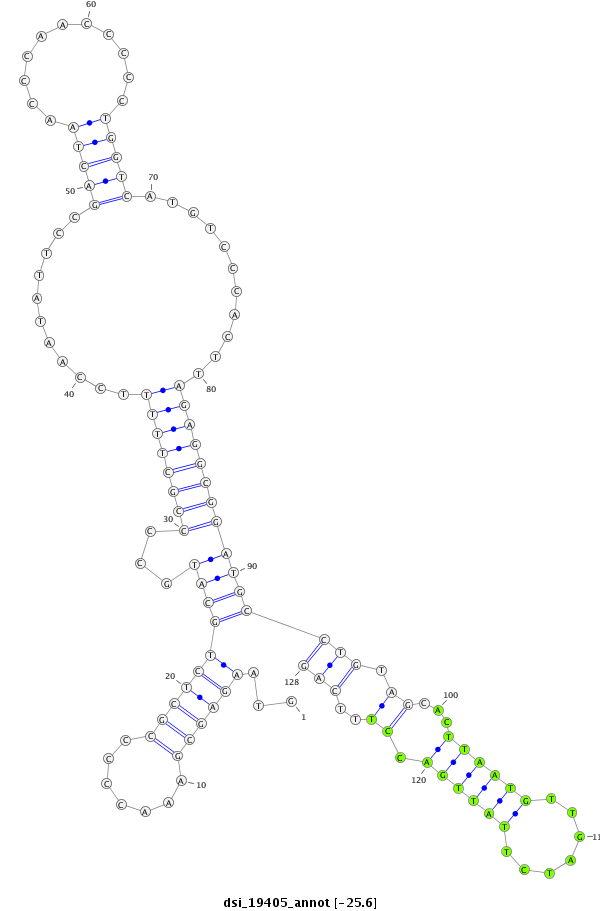
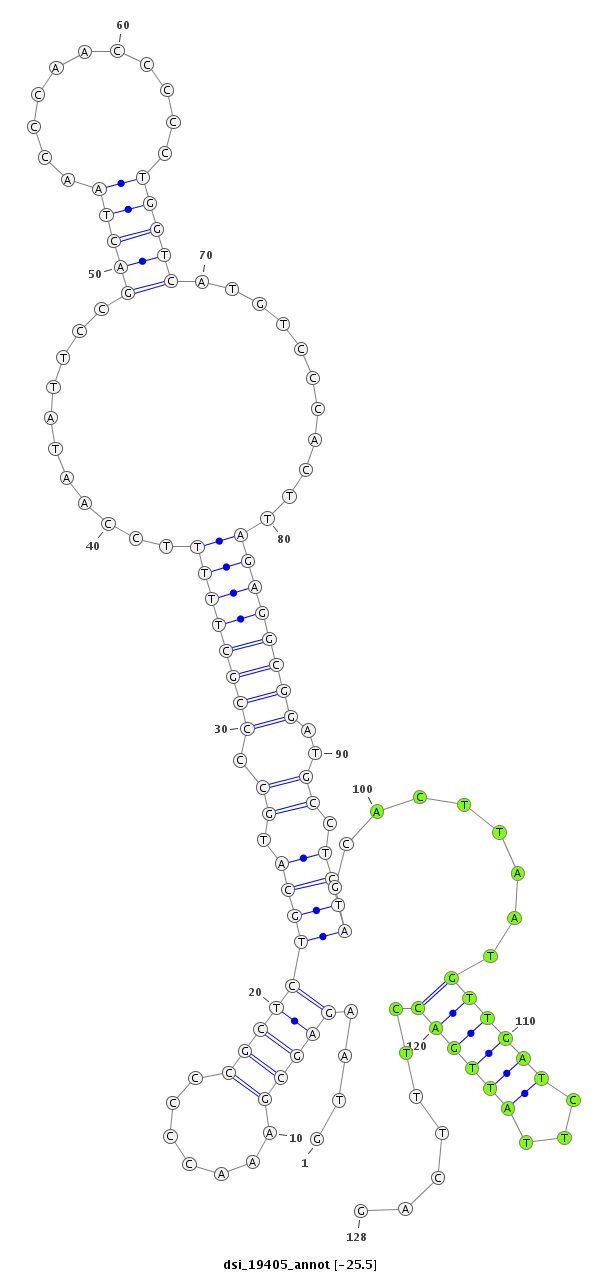
| -25.6 | -25.5 | -25.5 |
 |
 |
 |
exon [2r_4783693_4783797_+]; CDS [2r_4783693_4783797_+]; CDS [2r_4783926_4784026_+]; exon [2r_4783926_4784026_+]; intron [2r_4783798_4783925_+]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------################################################## GCCAAGCTAAAGGATGCGACCACGGCCAGTAGCGCCGCTCACTACGACAGGTAAGAGCGAAACCCCCGCTCTGCATGCCCCGCTTTTTCCAATATTCCGACTAACCCAACCCCCTGGTCATGTCCCACTTAGAGGCGGATGCCTGTAGCACTTAATGTTGATCTTATTGACCTTTCAGTGTCTCGACCAGCAGCGGAGTGAGCAGCAACAGCGGAAGCACAGGCAATG **************************************************(((((((((.......))).(((((.((.((((((((...........(((((...........)))))...........))))))))..)).))))).............))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553486 Makindu_3 day-old ovaries |
M023 head |
O001 Testis |
SRR553485 Chicharo_3 day-old ovaries |
SRR553487 NRT_0-2 hours eggs |
SRR618934 dsim w501 ovaries |
SRR553488 RT_0-2 hours eggs |
M025 embryo |
O002 Head |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .CCAAGCTAAAGGATGCGAC................................................................................................................................................................................................................ | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................GCGGAGTGAGCAGCAACAGC................ | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................GTCTAGACCCGAAGCGGAGTG............................ | 21 | 3 | 2 | 1.50 | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TCTGGACCCGAAGCGGAGTGA........................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AGCGGAAGCACAGGCA... | 16 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AGCAGCGGAGTGAGCAGCAATGGA................ | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................CGCCGCTCATTACGACAG.................................................................................................................................................................................. | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ACCAGCAGCGGAGTGAGCAGCAAC................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................................................................................CAGCGGAGTGAGCAGCAAC................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................ACTTAATGTTGATCTTATTGACCT....................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................TGCCTGTAGCACTTAATGTTGATCTT............................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................ACAGTGTCTCGACCAGCAGCGGAG.............................. | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................TAACCCAACCGCGTGGTC............................................................................................................. | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................................................GACCCGAAGCGGAGTGA........................... | 17 | 2 | 9 | 0.22 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................CGGCCTGTAGCGCAGCT............................................................................................................................................................................................. | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................ACCCGCAGCGGCGTGAG.......................... | 17 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................CCCTGTAGGACTTAATGCT..................................................................... | 19 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TTAGCGGCGGATGCGTG................................................................................... | 17 | 2 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ......................CGGCCTGTAGCGCAGC.............................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................AACGGCCGGTAGAGCCGC.............................................................................................................................................................................................. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GCATCAGCGGAAGCA......... | 15 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CGGTTCGATTTCCTACGCTGGTGCCGGTCATCGCGGCGAGTGATGCTGTCCATTCTCGCTTTGGGGGCGAGACGTACGGGGCGAAAAAGGTTATAAGGCTGATTGGGTTGGGGGACCAGTACAGGGTGAATCTCCGCCTACGGACATCGTGAATTACAACTAGAATAACTGGAAAGTCACAGAGCTGGTCGTCGCCTCACTCGTCGTTGTCGCCTTCGTGTCCGTTAC
**************************************************(((((((((.......))).(((((.((.((((((((...........(((((...........)))))...........))))))))..)).))))).............))))))...........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M023 head |
SRR618934 dsim w501 ovaries |
M024 male body |
SRR553488 RT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................TGGAAAGTCACAGAGCTGGT....................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................CGGAAAGTCACAGAGCTGGT....................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AGAGCTGGTCGTTGCC................................ | 16 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................GTCGTTGTCACCTTCGGGTC...... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................CAGAGCTGGTCGTTG.................................. | 15 | 1 | 10 | 0.30 | 3 | 0 | 3 | 0 | 0 |
| ..................................................................................................................................................................................GCAGAGCTGGTCGTTGCC................................ | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................AGAGCTGGTCGTTGC................................. | 15 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................AAAAAGGTTATAAGGTT................................................................................................................................ | 17 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................AGGTTATAAGGGTGA.............................................................................................................................. | 15 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................CAGAGCTGGTCGTTGCCAT.............................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................AAAAGGTTATCGGGCGGAT............................................................................................................................. | 19 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................CTGGTCGACGCCTCA............................. | 15 | 1 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ......................................CGTGCTGCTGTCCATT.............................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................AGAGCTGGTCGTTGCCA............................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
Generated: 04/23/2015 at 03:51 AM