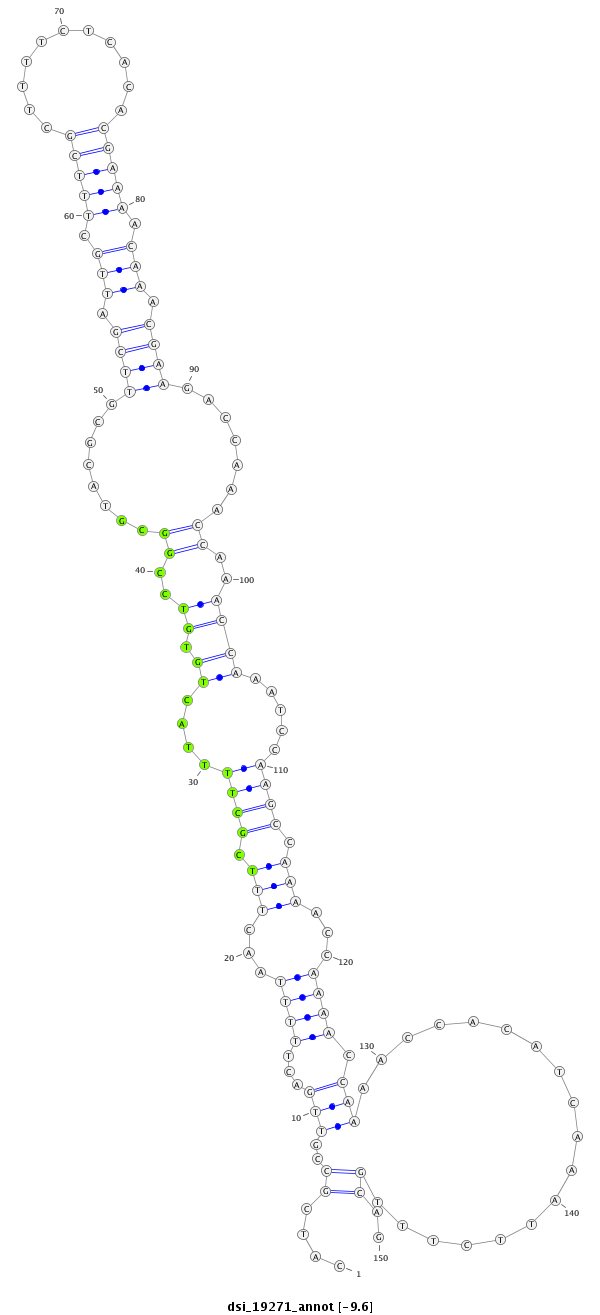
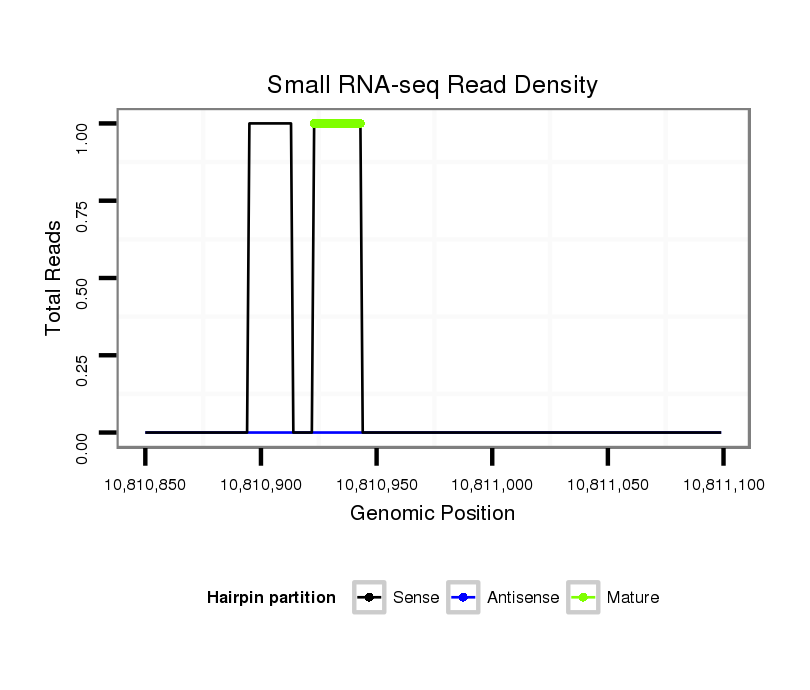
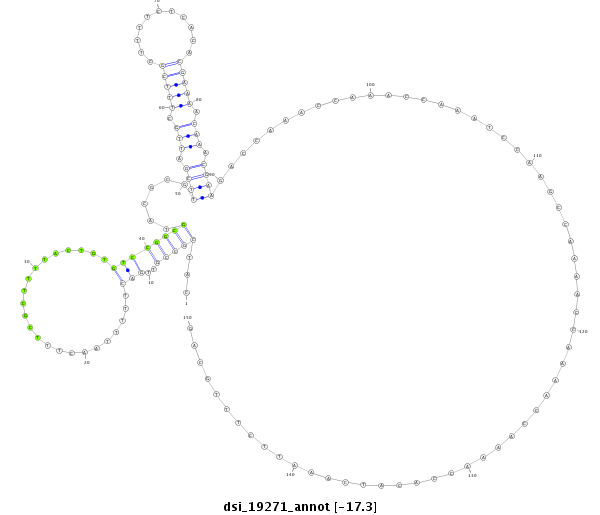
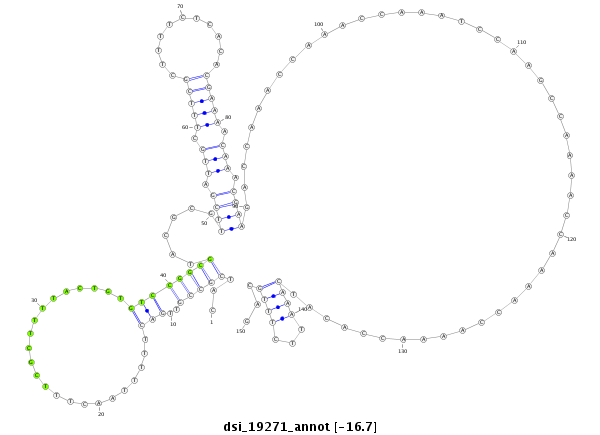
| CAGGCCGATCCTGCTTCTTC--TTCTTCTGTCCGTCCCGCACG------TTG--TTGCTG----------CTGCTGTTG------------GT----GCTG---------------CTGCCGTTGACTT-TTAACTTTCGCTTTTACT---------GTGT-------CG-----GCGTCCCCGCGACGCGCCACCG------CCACCGCC----CCGCCGC-TTACTTTCGTT-T-----------TCTCACA--CC----------------------ACAC-----------------------------------------------------------------------------------------------------------------ACAAAATTCTTTGCAGCG--CGCCAAAAATA--AGAACAGAACTCTG-TTTT-AGA------ACAC---TCGTA------------------------------------------ATCGCAC | Size | Hit Count | Total Norm | Total | M021
Embryo | M042
Female-body | V042
Embryo | V050
Head | V057
Head | V111
Male-body |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................ATA--AGAACAGAACTCTG-T.......................................................................... | 18 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AAAAATA--AGAACAGAACTCTG-T.......................................................................... | 22 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CGCCAAAAATA--AGAACAGAACTCTG-T.......................................................................... | 26 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................AATA--AGAACAGAACTCTG-T.......................................................................... | 19 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................TTCGCTTTTACT---------GTGT-------CG-----GC....................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAGAACTCTG-TTTT-AGA------ACAC---TC.................................................... | 24 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................CCAAAAATA--AGAACAGAACTCTG-T.......................................................................... | 24 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................................GAACAGAACTCTG-TTTT-AGA------ACAC---T..................................................... | 25 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................AAATA--AGAACAGAACTCTG-T.......................................................................... | 20 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................CAAAAATA--AGAACAGAACTCTG-T.......................................................................... | 23 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................TTT-AGA------ACAC---TCGTA------------------------------------------ATCGCAC | 22 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AAAAATA--AGAACAGAACTCT............................................................................. | 20 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................TTACT---------GTGT-------CG-----GCGTCCCC................................................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................AAATA--AGAACAGAACTCT............................................................................. | 18 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CGCCAAAAATA--AGAACAGAACTCTG-G.......................................................................... | 26 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CGCCAAAAATA--AGAACAGAACTCTG-........................................................................... | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................TT-TTAACTTTCGCTTTTACT---------GTGT-------C............................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGAACTCTG-TTTT-AGA------AC........................................................... | 19 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................GTCCCGCACG------TTG--TTGCTG----------CTGC.............................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................AACTTTCGCTTTTACT---------GTGT-------C............................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................AAAATA--AGAACAGAACTCTG-T.......................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................AAATA--AGAACAGAACTCTG-G.......................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................ACAGAACTCTG-TTTT-AGA------ACAC---T..................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......GATCCTGCTTCTTC--TTCTTCTGG......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................GTTGACTT-TTAACTTTCGCT.......................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................CTTTCGCTTTTACT---------GTGT-------CG-----GC....................................................................................................................................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................AACAGAACTCTG-TTTT-AGA------ACAC---TC.................................................... | 25 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................CGCCAAAAATA--AGAACAGAACTCT............................................................................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................AATA--AGAACAGAACTCTG-C.......................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................TGCCGTTGACTT-TTAACTTTCGCT.......................................................................................................................................................................................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................CTCTG-TTTT-AGA------ACAC---TCGTA------------------------------------------ATCGCAC | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................CAAAAATA--AGAACAGAACTCTG-TT......................................................................... | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................CGCTTTTACT---------GTGT-------CG-----GC....................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................CAAAAATA--AGAACAGAACTCG............................................................................. | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AAAAATA--AGAACAGAACTCTG-G.......................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................CAGAACTCTG-TTTT-AGA------ACAC---T..................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................AAAATA--AGAACAGAACTCT............................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................G-TTTT-AGA------ACAC---TCGTA------------------------------------------ATCGCAC | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................AATA--AGAACAGAACTCTG-TTTT-...................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AAAAATA--AGAACAGAACTCTG-C.......................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................AATA--AGAACAGAACTCTG-G.......................................................................... | 19 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |