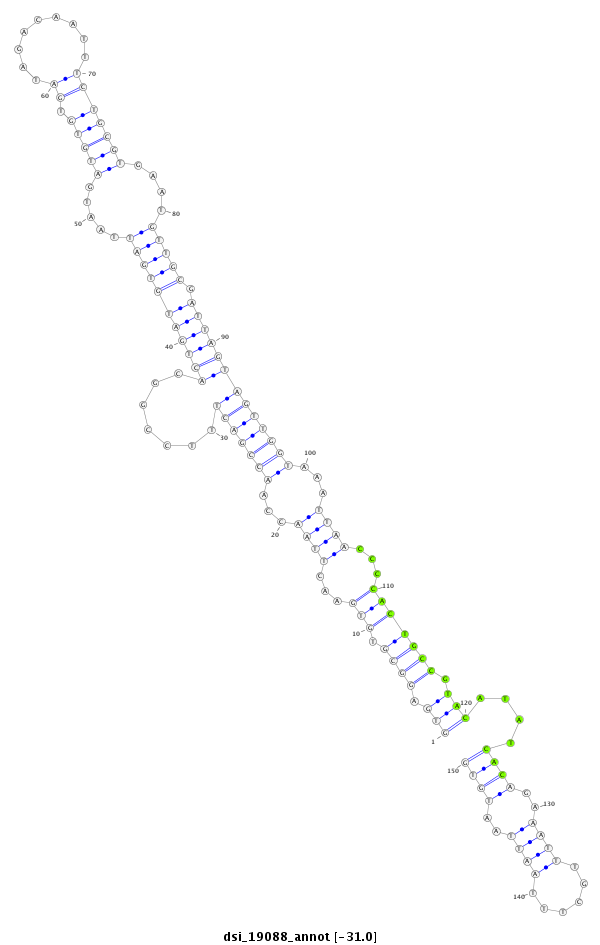
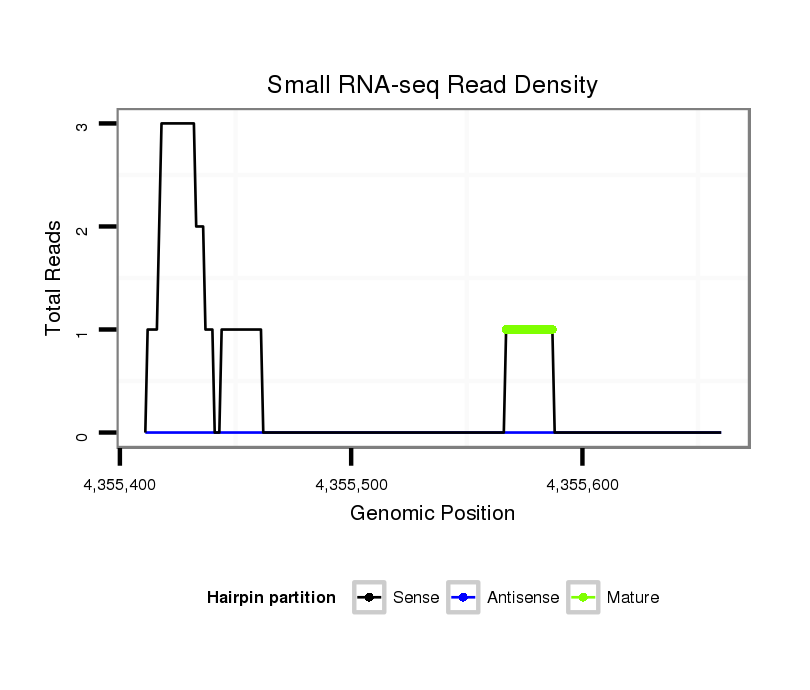

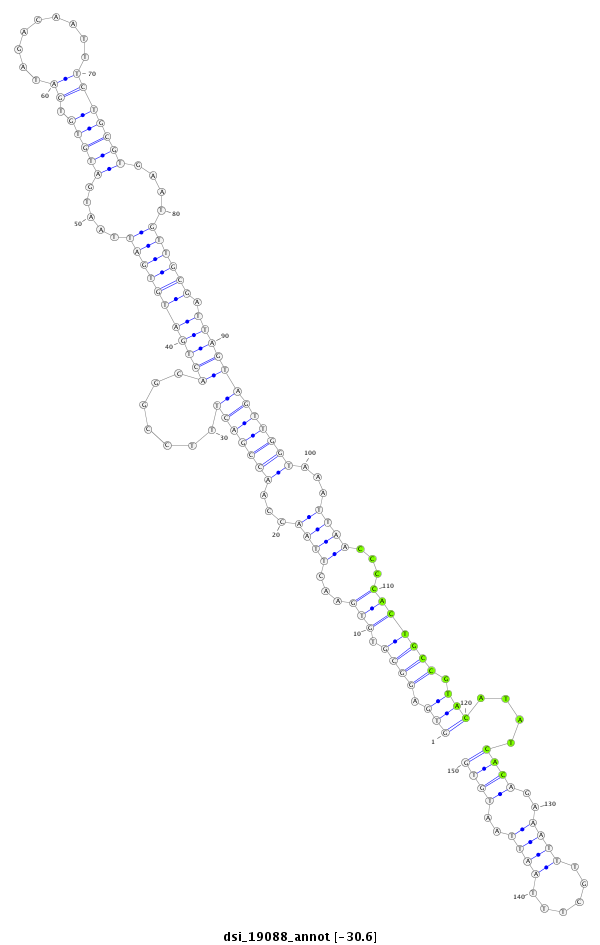
ID:dsi_19088 |
Coordinate:3r:4355461-4355610 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in read |
 |
 |
 |
 |
| -30.8 | -30.6 | -30.6 |
 |
 |
 |
exon [3r_4355006_4355460_+]; CDS [3r_4355006_4355460_+]; intron [3r_4355461_4355776_+]
No Repeatable elements found
| ##################################################-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAGTACAGAGATCGCCTGGTGGTCAGCGGCAGCTCAGACAATTCCATAAGGTGAGGCGTGTGAACTTAACCAACCGACTTTCCGGCACTGATGTGATTAATGATGTGTGATAGACAATTTCTGCGTGAATGTTGCGATTAGTAGTTGGTAAATTAACCCCACTGCCGTACATATCACAGAAATTTGCTTTAATTAATGTGTATTTCCTTGCTTTTTGTTAGCATCTGAATATTAAATATTATATTAAGAT **************************************************(((.((((.(((...((((...(((((((.......(((((((((((.....(((((.((.........)))))))....))))).)))))))))))))...))))...))))))).)))....((((..((((......))))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR618934 dsim w501 ovaries |
SRR553485 Chicharo_3 day-old ovaries |
O001 Testis |
SRR553486 Makindu_3 day-old ovaries |
GSM343915 embryo |
SRR553487 NRT_0-2 hours eggs |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .......GAGATCGCCTGGTGG.................................................................................................................................................................................................................................... | 15 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................CCCCACTGCCGTACATATCAC......................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .................................TCAGACAATTCCATAAGG....................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......AGAGATCGCCTGGTGGTCAGCGGC............................................................................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................AATGATGCGATTAGTAGTCG....................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .AGTACAGAGATCGCCTGGTGGTCAG................................................................................................................................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ........AGAACGCCTGGTGGT................................................................................................................................................................................................................................... | 15 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................................TTAACGTGTATTTCCT.......................................... | 16 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................TATCACAGAGTTTTGCTT............................................................. | 18 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................AAGGAGAATCGTGTGAACT........................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................AATGTTGGCATTAGTAG.......................................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........GAACGCCTGGTGGTTA................................................................................................................................................................................................................................. | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
GTCATGTCTCTAGCGGACCACCAGTCGCCGTCGAGTCTGTTAAGGTATTCCACTCCGCACACTTGAATTGGTTGGCTGAAAGGCCGTGACTACACTAATTACTACACACTATCTGTTAAAGACGCACTTACAACGCTAATCATCAACCATTTAATTGGGGTGACGGCATGTATAGTGTCTTTAAACGAAATTAATTACACATAAAGGAACGAAAAACAATCGTAGACTTATAATTTATAATATAATTCTA
**************************************************(((.((((.(((...((((...(((((((.......(((((((((((.....(((((.((.........)))))))....))))).)))))))))))))...))))...))))))).)))....((((..((((......))))..))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | SRR553487 NRT_0-2 hours eggs |
GSM343915 embryo |
SRR553488 RT_0-2 hours eggs |
M024 male body |
|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................ATTACTACACACTAGCTCTT..................................................................................................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................ACAGCCATTCGCCGTCGAGT...................................................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................ATTGGTTGGCGGAATGGCA..................................................................................................................................................................... | 19 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................GGGTGTTGGGATGTATAGT.......................................................................... | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................TCCAATGGGTTGGCTGAAA......................................................................................................................................................................... | 19 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 |
| ....TGTCTGTAGGGGACCA...................................................................................................................................................................................................................................... | 16 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
Generated: 04/24/2015 at 02:51 AM